
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Serge Rozov | -- | 2717 | 2022-04-25 13:41:46 | | | |
| 2 | Beatrix Zheng | -1 word(s) | 2716 | 2022-04-26 04:34:32 | | |
Video Upload Options
Plant expression systems are currently regarded as promising alternative platforms for the production of recombinant proteins, including the proteins for biopharmaceutical purposes. However, the accumulation level of a target protein in plant expression systems is still rather low compared with the other existing systems, namely, mammalian, yeast, and E. coli cells. To solve this problem, numerous methods and approaches have been designed and developed. At the same time, the random nature of the distribution of transgenes over the genome can lead to gene silencing, variability in the accumulation of recombinant protein, and also to various insertional mutations. The current research considered inserting target genes into pre-selected regions of the plant genome (genomic “safe harbors”) using the CRISPR/Cas system. Regions of genes expressed constitutively and at a high transcriptional level in plant cells (housekeeping genes) that are of interest as attractive targets for the delivery of target genes were characterized.
1. Introduction
2. Activation (Euchromatization) of Certain Genomic Regions
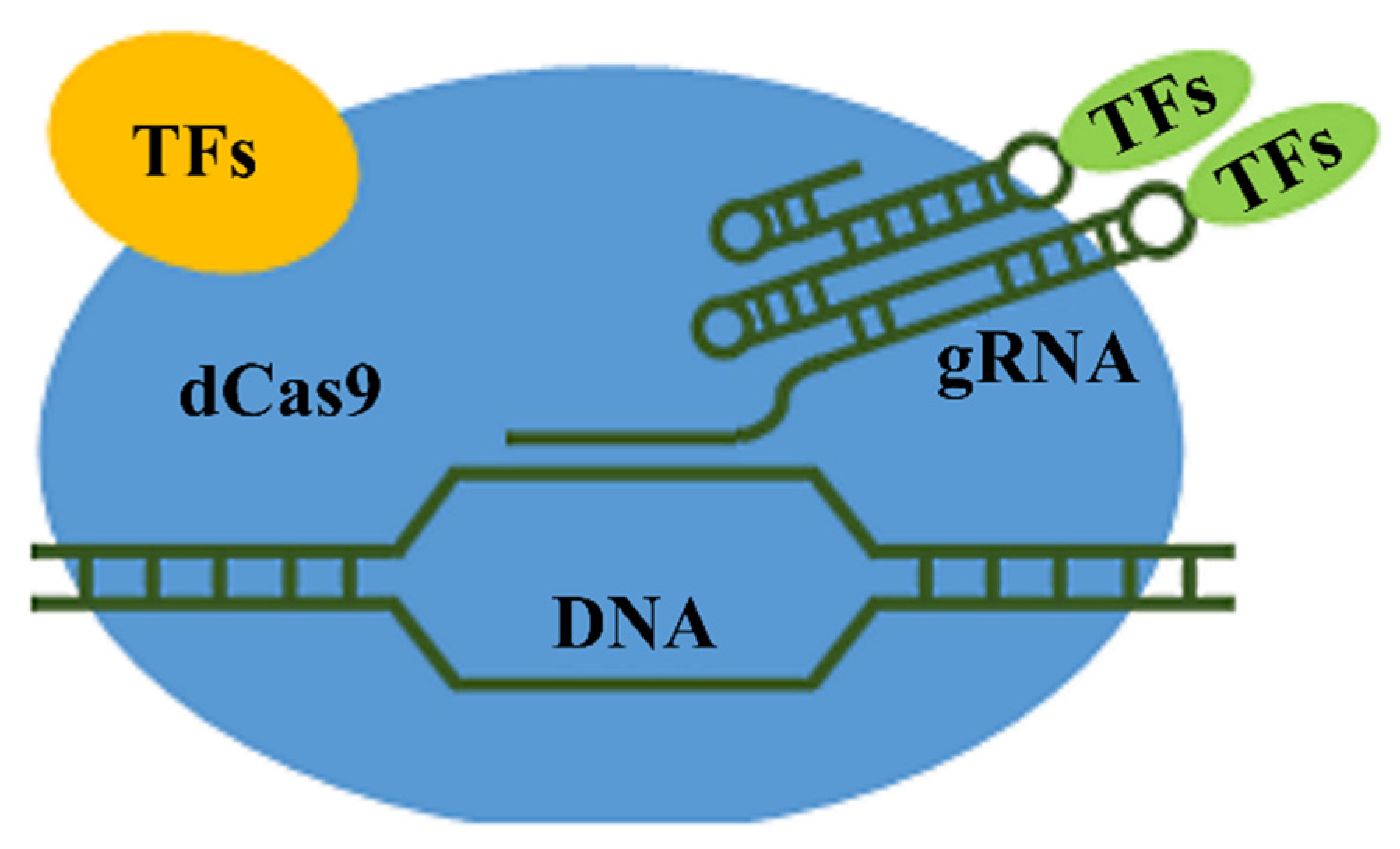
3. Spatial Organization of Chromatin and Transcriptional Activity
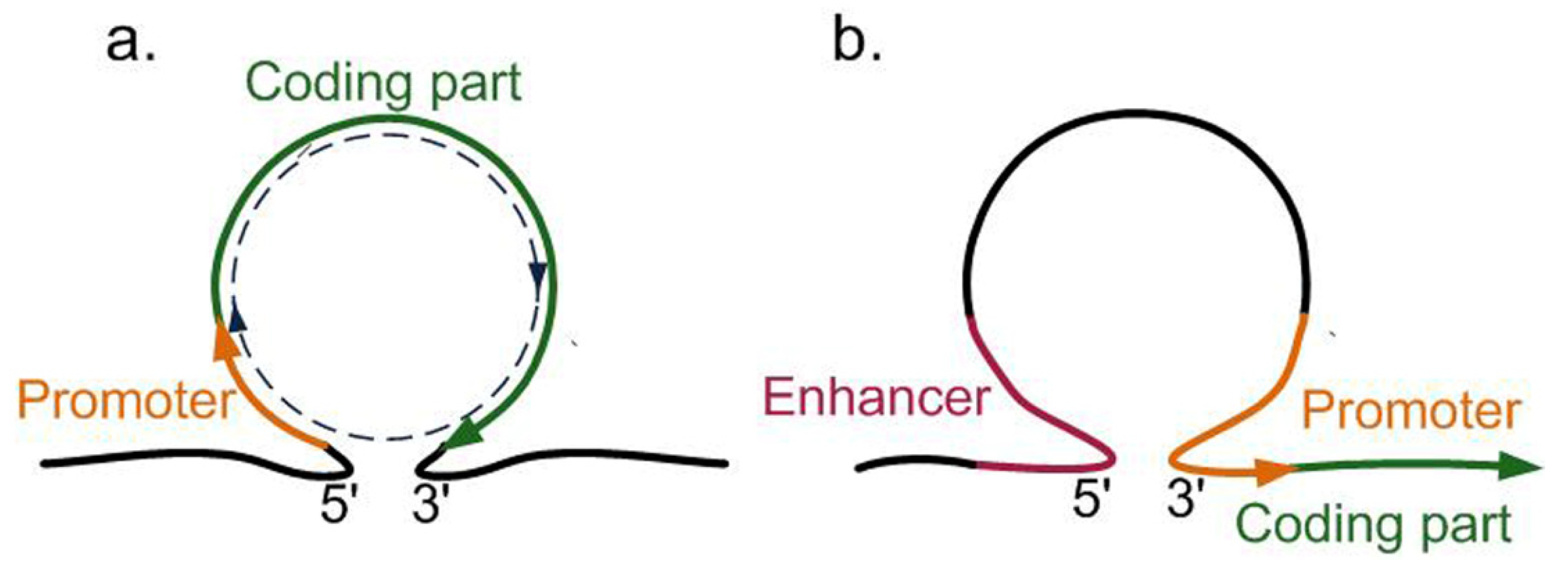
4. Conclusions
References
- Burnett, M.J.B.; Burnett, A.C. Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants People Planet 2020, 2, 121–132.
- Huebbers, J.W.; Buyel, J.F. On the verge of the market—Plant factories for the automated and standardized production of biopharmaceuticals. Biotechnol. Adv. 2021, 46, 107681.
- Schillberg, S.; Finnern, R. Plant molecular farming for the production of valuable proteins—Critical evaluation of achievements and future challenges. J. Plant Physiol. 2021, 258–259, 153359.
- Karki, U.; Fang, H.; Guo, W.; Unnold-Cofre, C.; Xu, J. Cellular engineering of plant cells for improved therapeutic protein production. Plant Cell Rep. 2021, 40, 1087–1099.
- Rozov, S.M.; Deineko, E.V. Strategies for Optimizing Recombinant Protein Synthesis in Plant Cells: Classical Approaches and New Directions. Mol. Biol. 2019, 53, 179–199.
- Buyel, J.F.; Stöger, E.; Bortesi, L. Targeted genome editing of plants and plant cells for biomanufacturing. Transgenic Res. 2021, 30, 401–426.
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.P.; Tuteja, N. The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front. Plant Sci. 2016, 7, 506.
- Demirci, Y.; Zhang, B.; Unver, T. CRISPR/Cas9: An RNA-guided highly precise synthetic tool for plant genome editing. J. Cell. Physiol. 2018, 233, 1844–1859.
- Huang, T.K.; Puchta, H. CRISPR/Cas-mediated gene targeting in plants: Finally a turn for the better for homologous recombination. Plant Cell Rep. 2019, 38, 443–453.
- Rozov, S.M.; Permyakova, N.V.; Deineko, E.V. The Problem of the Low Rates of CRISPR/Cas9-Mediated Knock-ins in Plants: Approaches and Solutions. Int. J. Mol. Sci. 2019, 20, 3371.
- Collonnier, C.; Guyon-Debast, A.; Maclot, F.; Mara, K.; Charlot, F.; Nogué, F. Towards mastering CRISPR-induced gene knock-in in plants: Survey of key features and focus on the model Physcomitrella patens. Methods 2017, 121–122, 103–117.
- Dong, O.X.; Ronald, P.C. Targeted DNA insertion in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2004834117.
- Rebetzke, G.J.; Jimenez-Berni, J.; Fischer, R.A.; Deery, D.M.; Smith, D.J. Review: High-throughput phenotyping to enhance the use of crop genetic resources. Plant Sci. 2019, 282, 40–48.
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 187–214.
- Dong, O.X.; Yu, S.; Jain, R.; Zhang, N.; Duong, P.Q.; Butler, C.; Li, Y.; Lipzen, A.; Martin, J.A.; Barry, K.W.; et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat. Commun. 2020, 11, 1–10.
- Permyakova, N.V.; Marenkova, T.V.; Belavin, P.A.; Zagorskaya, A.A.; Sidorchuk, Y.V.; Uvarova, E.A.; Kuznetsov, V.V.; Rozov, S.M.; Deineko, E.V. Assessment of the Level of Accumulation of the dIFN Protein Integrated by the Knock-In Method into the Region. Cells 2021, 10, 2137.
- Belavin, P.A.; Permyakova, N.V.; Zagorskaya, A.A.; Marenkova, T.V.; Sidorchuk, Y.V.; Uvarova, E.A.; Rozov, S.M.; Deineko, E.V. Peculiarities in Creation of Genetic Engineering Constructions for Knock-In Variant of Genome Editing of Arabidopsis thaliana Cell Culture. Russ. J. Plant Physiol. 2020, 67, 855–866.
- Joseph, J.T.; Poolakkalody, N.J.; Shah, J.M. Plant reference genes for development and stress response studies. J. Biosci. 2018, 43, 173–187.
- Maeder, M.L.; Linder, S.J.; Cascio, V.M.; Fu, Y.; Ho, Q.H.; Joung, J.K. CRISPR RNA—Guided activation of endogenous human genes. Nat. Methods 2013, 10, 977.
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR- Cas9—Based transcription factors. Nat. Methods 2013, 10, 973.
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015, 169, 971–985.
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 2015, 13, 578–589.
- Papikian, A.; Liu, W.; Gallego-Bartolomé, J.; Jacobsen, S.E. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 2019, 10, 1–11.
- Xiong, X.; Liang, J.; Li, Z.; Gong, B.Q.; Li, J.F. Multiplex and optimization of dCas9-TV-mediated gene activation in plants. J. Integr. Plant Biol. 2021, 63, 634–645.
- Selma, S.; Bernabé-Orts, J.M.; Vazquez-Vilar, M.; Diego-Martin, B.; Ajenjo, M.; Garcia-Carpintero, V.; Granell, A.; Orzaez, D. Strong gene activation in plants with genome-wide specificity using a new orthogonal CRISPR/Cas9-based programmable transcriptional activator. Plant Biotechnol. J. 2019, 17, 1703–1705.
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.F.; Voytas, D.F.; Zhang, Y.; Qi, Y. Robust Transcriptional Activation in Plants Using Multiplexed CRISPR-Act2.0 and mTALE-Act Systems. Mol. Plant 2018, 11, 245–256.
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 2021, 7, 942–953.
- Doğan, E.S.; Liu, C. Three-dimensional chromatin packing and positioning of plant genomes. Nat. Plants 2018, 4, 521–529.
- Stam, M.; Tark-Dame, M.; Fransz, P. 3D genome organization: A role for phase separation and loop extrusion? Curr. Opin. Plant Biol. 2019, 48, 36–46.
- Gibcus, J.H.; Dekker, J. The Hierarchy of the 3D Genome. Mol. Cell 2013, 49, 773–782.
- Liu, C.; Wang, C.; Wang, G.; Becker, C.; Zaidem, M.; Weigel, D. Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res. 2016, 26, 1057–1068.
- Dekker, J.; Marti-Renom, M.A.; Mirny, L.A. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat. Rev. Genet. 2013, 14, 390–403.
- Chung, I.M.; Ketharnathan, S.; Kim, S.H.; Thiruvengadam, M.; Rani, M.K.; Rajakumar, G. Making sense of the tangle: Insights into chromatin folding and gene regulation. Genes 2016, 7, 71.
- Rodriguez-Granados, N.Y.; Ramirez-Prado, J.S.; Veluchamy, A.; Latrasse, D.; Raynaud, C.; Crespi, M.; Ariel, F.; Benhamed, M. Put your 3D glasses on: Plant chromatin is on show. J. Exp. Bot. 2016, 67, 3205–3221.
- Pontvianne, F.; Liu, C. Chromatin domains in space and their functional implications. Curr. Opin. Plant Biol. 2020, 54, 1–10.
- Gagliardi, D.; Manavella, P.A. Short-range regulatory chromatin loops in plants. New Phytol. 2020, 228, 466–471.
- Larkin, J.D.; Cook, P.R.; Papantonis, A. Dynamic Reconfiguration of Long Human Genes during One Transcription Cycle. Mol. Cell. Biol. 2012, 32, 2738–2747.
- Cavalli, G.; Misteli, T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 2013, 20, 290–299.
- Gagliardi, D.; Cambiagno, D.A.; Arce, A.L.; Tomassi, A.H.; Giacomelli, J.I.; Ariel, F.D.; Manavella, P.A. Dynamic regulation of chromatin topology and transcription by inverted repeat-derived small RNAs in sunflower. Proc. Natl. Acad. Sci. USA 2019, 116, 17578–17583.
- Crevillén, P.; Sonmez, C.; Wu, Z.; Dean, C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013, 32, 140–148.
- Jégu, T.; Domenichini, S.; Blein, T.; Ariel, F.; Christ, A.; Kim, S.K.; Crespi, M.; Boutet-Mercey, S.; Mouille, G.; Bourge, M.; et al. A SWI/SNF chromatin remodelling protein controls cytokinin production through the regulation of chromatin architecture. PLoS ONE 2015, 10, e0138276.
- Louwers, M.; Bader, R.; Haring, M.; Van Driel, R.; De Laat, W.; Stam, M. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell 2009, 21, 832–842.
- Cao, S.; Kumimoto, R.W.; Gnesutta, N.; Calogero, A.M.; Mantovani, R.; Holt, B.F. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 2014, 26, 1009–1017.
- Guo, L.; Cao, X.; Liu, Y.; Li, J.; Li, Y.; Li, D.; Zhang, K.; Gao, C.; Dong, A.; Liu, X. A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J. 2018, 94, 1083–1097.
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding transcription by alternative rna polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 2014, 55, 383–396.
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4.




