Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Elisabeth Jamet | -- | 3636 | 2022-04-20 12:35:44 | | | |
| 2 | Amina Yu | -87 word(s) | 3549 | 2022-04-21 04:13:48 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Jamet, E.; San Clemente, H.; , .; Canut, H. Plant Cell Wall Proteomes. Encyclopedia. Available online: https://encyclopedia.pub/entry/22008 (accessed on 07 February 2026).
Jamet E, San Clemente H, , Canut H. Plant Cell Wall Proteomes. Encyclopedia. Available at: https://encyclopedia.pub/entry/22008. Accessed February 07, 2026.
Jamet, Elisabeth, Helene San Clemente, , Hervé Canut. "Plant Cell Wall Proteomes" Encyclopedia, https://encyclopedia.pub/entry/22008 (accessed February 07, 2026).
Jamet, E., San Clemente, H., , ., & Canut, H. (2022, April 20). Plant Cell Wall Proteomes. In Encyclopedia. https://encyclopedia.pub/entry/22008
Jamet, Elisabeth, et al. "Plant Cell Wall Proteomes." Encyclopedia. Web. 20 April, 2022.
Copy Citation
Cell wall proteins (CWPs) play critical roles in the biogenesis of plant cell walls and in their rearrangement during plant growth and development as well as in response to environmental constraints. Many cell wall proteomes have now been described, thus allowing drawing a general picture.
cell wall
cell wall protein
early divergent plant
flowering plant
green lineage
proteomics
non-canonical extracellular protein
1. A Core Cell Wall Proteome: The Conserved CWPs Families and Their Possible Roles in Cell Walls
Thirteen cell wall proteomes have been selected for this survey: Marchantia polymorpha as an early divergent plant, Brachypodium distachyon, Oryza sativa, Triticum aestivum and Saccharum officinarum as monocotyledonous plants, and Arabidopsis thaliana, Linum usitatissimum, Medicago sativa, Populus spp, Solanum lycopersicum, Solanum tuberosum, Gossypium hirsutum, and Camellia sinensis as dicotyledonous plants. These proteomes were chosen because they were obtained in similar conditions, they had a minimal size of 100 proteins and the available data have allowed a new expert annotation thanks to bioinformatics tools and data. The identified proteins were sorted into cell wall proteins (CWPs) and presumed intracellular contaminants.
The systematic re-annotation of the CWPs after the presence of predicted signal peptide and functional domains has allowed grouping them into nine functional classes [1], which have been found in various proportions in the cell wall proteomes of the 13 studied plant species:
-
Proteins acting on cell wall carbohydrates (PACs) belong to the major functional class in all the cell wall proteomes accounting for up to 25% of the CWPs. It comprises expansins [2] as well as glycosyl hydrolases (GHs), carbohydrate esterases (CEs) such as pectin methylesterases (PMEs) and polysaccharide lyases (PLs). The description of the latter protein families can be found in the Carbohydrate-Active enZYmes Database (CAZyDB, http://www.cazy.org) [3].
-
Oxido-reductases (ORs) include class III peroxidases (CIII Prxs), blue copper binding proteins, berberine bridge oxido-reductases (BBEs), multicopper oxidases and laccases. The CIII Prxs and blue copper binding proteins are described in the Redoxibase (https://peroxibase.toulouse.inrae.fr) [4] and the two latter protein families are included in CAZyDB.
-
Proteins with interaction domains comprise proteins interacting with other proteins, such as enzyme inhibitors, or with cell wall carbohydrates, such as lectins [13].
-
Structural proteins, such as hydroxyproline-rich glycoproteins (HRGPs), are scarcely represented in cell wall proteomes because many of them are covalently cross-linked in cell walls like extensins, and thus difficult to extract, or because they behave as proteoglycans, like AGPs. It has particularly succeeded in the identification of several extensins, Pro-rich proteins (PRPs) and leucine-rich extensins (LRXs) by using a dedicated protocol including a trypsin digestion applied directly on cell walls [16].
-
Proteins of unknown function can represent more than one tenth of the cell wall proteomes, suggesting new functions or new biological activities yet to be described.
As mentioned, each of these functional classes includes several protein families. By comparing the 13 selected cell wall proteomes, it is possible to identify protein families which are present in all or in most of them . They are described in the two following paragraphs: proteins acting on cell wall carbohydrates belonging to the major functional class (Figure 1) and proteins belonging to the other functional classes (Figure 2).
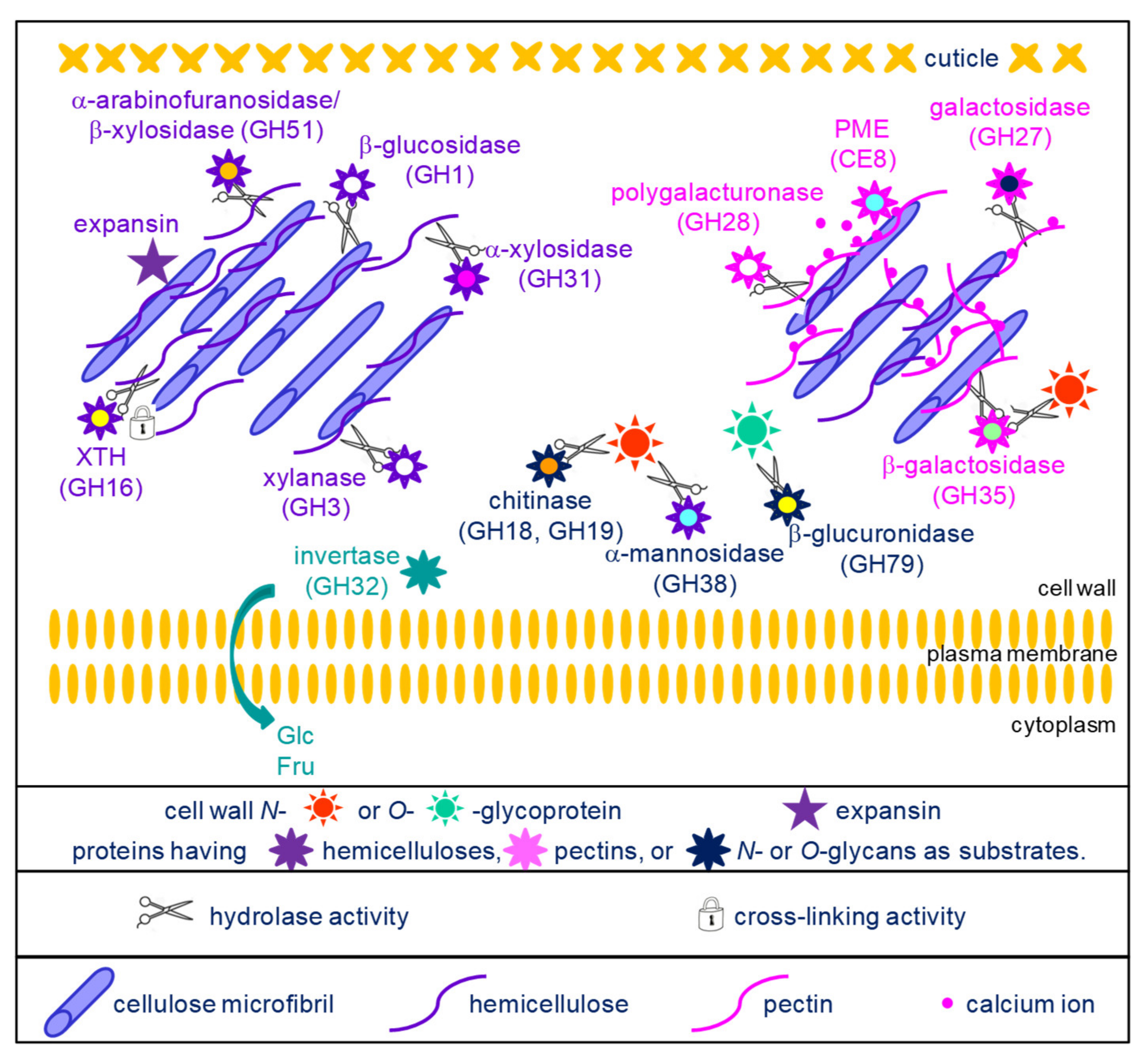
Figure 1. Schematic representation of the activities of the proteins acting on cell wall carbohydrates and belonging to the core cell wall proteome. With the exception of expansins and CE8, all the protein families are glycoside hydrolases (GHs) which were grouped according to their possible substrates: cellulose and hemicelluloses for GH1, GH3, GH16, GH31, GH51 and expansins (top left part of the scheme); pectins for GH27, GH28, GH35 and CE8 (top right part of the scheme); N- or O-glycans for GH18, GH19, GH38 and GH79 (center of the scheme); sucrose for GH32, thus releasing glucose (Glc) and fructose (Fru) which can be transferred to the cytoplasm by hexose transporters.
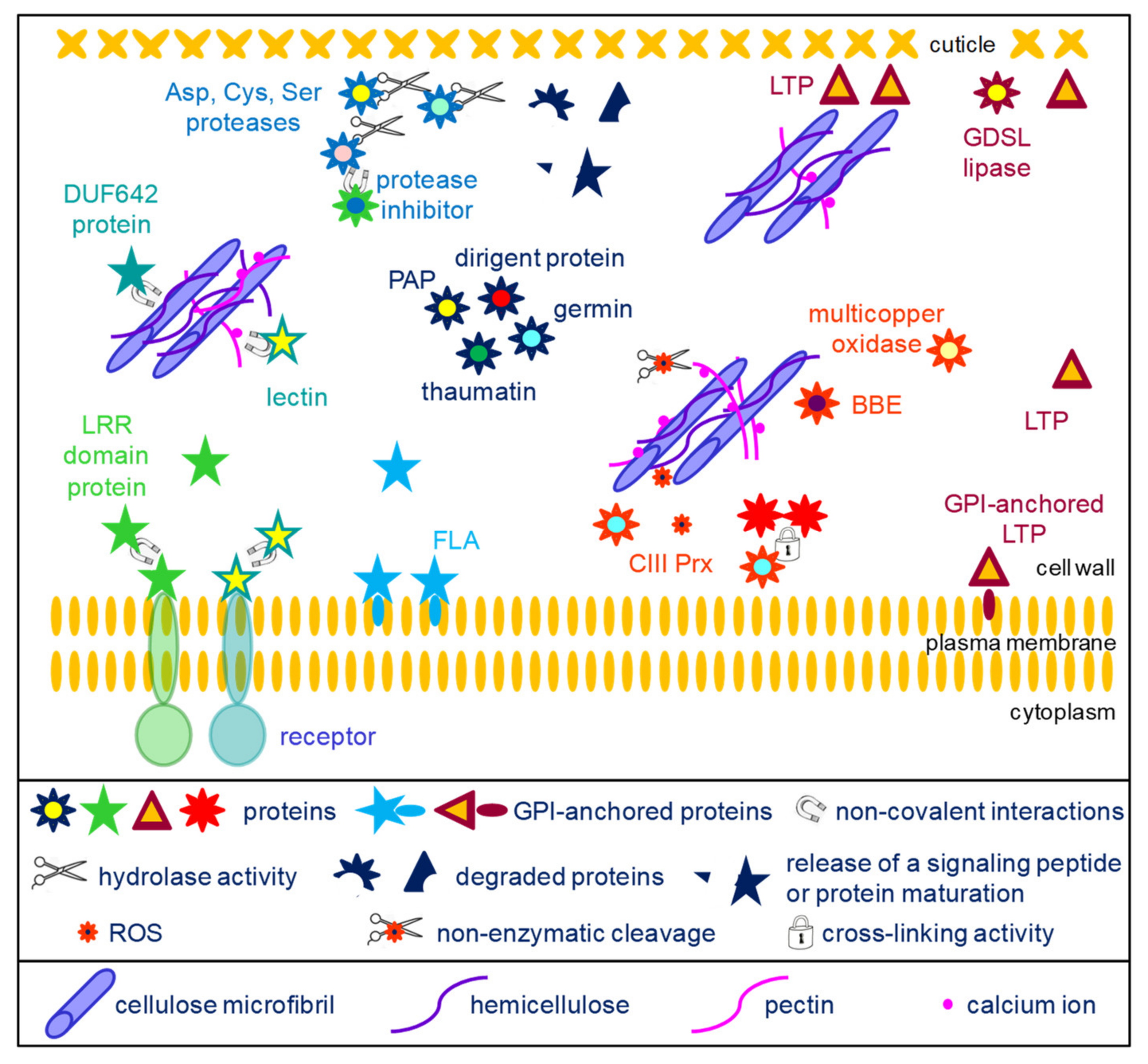
Figure 2. Schematic representation of the activities of diverse proteins belonging to the core cell wall proteome. The protein families have been grouped according to their known biological activities. Proteases are assumed to play roles in protein maturation, release of signaling peptides and protein degradation (top left of the scheme). DUF642 proteins and lectins interact with cell wall polysaccharides but their precise roles are not known (middle left part of the scheme). Several protein families could play roles in signaling (bottom left of the scheme): LRR proteins and lectins could interact with other proteins, and in particular with the extracellular domains of plasma membrane receptors, thus leading to the transduction of a signal to the cell; fasciclin arabinogalactan proteins (FLAs) are also assumed to play a role in signaling. Dirigent proteins, germins, thaumatins and purple acid phosphatases (PAPs) have diverse activities (center of the scheme). Oxido-reductases (multicopper oxidases, berberine-bridge oxido-reductases (BBEs) and class III peroxidases (CIII Prxs)) play multiple roles in the cell wall. In particular, CIII Prxs can cross-link structural proteins or phenolics compounds, and they contribute to the regulation of reactive oxygen species (ROS) which are involved in signaling or in non-enzymatic cleavage of polysaccharides (central part of the scheme). LTPs and GDSL lipases could play roles in the formation of cuticle (right side of the scheme). Some LTPs are localized at the surface of the plasma membrane thanks to GPI anchors and participate in the transport of lipids to the cuticle layer. LTPs have also been shown to play a role at the interface between the hydrophilic cell wall polysaccharides and the hydrophobic cuticle layer.
1.1. Proteins Acting on Cell Wall Carbohydrates
These protein families can be distinguished on the basis of their carbohydrate substrates. They have been grouped according to their known or predicted substrates: hemicelluloses, pectins or glycans of glycoproteins (Figure 1).
A set of enzymes can act on hemicelluloses. GH16 are xyloglucan endotransglucosylases/hydrolases (XTHs). They were initially described as having xyloglucan-xyloglucan donor/acceptor substrate activities. However, it was later shown that they could accept other substrates such as cellulose or mixed-linkage (1,3;1,4)-β-D-glucans [21][22][23]. Molecular modelling had suggested that they could also modify arabinoxylans in Poaceae [23]. These findings allow assuming that they could play critical roles in remodeling the cellulose/hemicellulose networks in cell walls of both monocot and dicot plants. As an example, the xth21 mutant of A. thaliana exhibited a dwarf phenotype most probably resulting from a defect in the growth of the primary root [24]. This mutant also showed a decrease in the average mass of xyloglucans and in cellulose content, suggesting the role of the cellulose/xyloglucan network in the elongation of the cell wall.
GH1 (mostly β-glucosidases), GH3 (xylanases), GH51 (arabinofuranosidases/β-xylosidases), GH31 (α-xylosidases) and GH17 (β-1,3-glucosidases) have a hydrolytic activity towards different types of hemicelluloses or callose [25]. A. thaliana mutants impaired in AtBG_ppap (β-1,3-glucanase_putative plasmodesmata-associated protein), have an increased amount of callose at the level of the plasmodesmata and the cell-to-cell movement of a fluorescent marker protein is slower than in wild type [26]. These results, together with identification of AtBG_ppap in a plasmodesmata proteome, suggest its role in the regulation of symplasmic communication.
Another group of enzymes can hydrolyze or modify pectin molecules. GH27 and GH28 hydrolyze galactomannans and homogalacturonans, respectively [25]. The A. thaliana QRT3 (QUARTET3) gene was shown to encode a polygalacturonase and the corresponding mutant exhibited defect in pollen mother cell wall degradation resulting in the defect in microspore separation [27]. GH35 could act on the arabinan side-chains of pectins or on the O-glycans of AGPs although some of them could also act on xyloglucans [25]. PMEs operate the demethylesterification of homogalacturonans, thus revealing negative charges which allow the formation of the egg box structures with calcium ions [28]. The A. thaliana atpme3 mutant was shown to have an increased number of adventitious roots together with an increase in the degree of HG methylesterification, thus suggesting the importance of changes in the pectin structure for adventitious root emergence [29].
Finally, a set of enzymes can hydrolyze the N- or the O-glycans of glycoproteins. They belong to GH families 18, 19 and 38 [30]. The O-glycans of AGPs were assumed to be substrates of GH19 as one of the few cell wall molecules carrying glucosamine or N-acetylglucosamine [31]. In the same one, it was shown that an incubation of an AGP fraction purified from carrot cells with an endochitinase of the GH19 family lead to the release of oligosaccharides. GH18 and GH19 were also described as chitinases/lysozymes playing roles during plant-microorganism interactions [32][33].
1.2. The Other Conserved Protein Families
Apart from the proteins acting on cell wall carbohydrates, several protein families are also conserved (Figure 2). Several families of extracellular proteases are well conserved in cell wall proteomes, such as Asp proteases, Cys proteases and Ser proteases. The roles of these proteins have begun to be discovered in A. thaliana. The AtSBT1.4, AtSBT1.7 and AtSBT4.13 subtilisins were shown to release the signaling peptide CLE40 (Clavata3/Endosperm Surrounding Region 40) from a preprotein [36]. CLE40 is involved in the regulation of stem cell differentiation. Such extracellular proteases may also play roles in protein maturation as AtSBT1.6 for PMEs [6]. The SDD1 (Stomatal Density and Distribution 1) subtilisin negatively regulates the formation of stomata in A. thaliana, most probably through peptide signaling, although its substrate has not yet been identified [37]. Besides, the A. thaliana extracellular CDR1 (Constitutive Disease Resistance) Asp protease was assumed to mediate disease resistance through a signaling peptide [38]. Most probably, all these proteolytic activities are modulated by proteases inhibitors which are also found as conserved protein families in cell walls.
Among the ORs, CIII Prxs represent large plant gene families, with, for example, 73 members in A. thaliana and 189 in M. polymorpha (https://peroxibase.toulouse.inrae.fr). They play major roles in plant cell walls by (i) generating reactive oxygen species (ROS) involved in signaling and in nonenzymatic cleavage of polysaccharides, or by regulating the level of H2O2, thus contributing to cell wall stiffening by cross-linking structural proteins such as extensins or monomers of lignins [39]. This latter role could also be played by laccases, such as LACCASE5 in B. distachyon culms [40]. Besides, an A. thaliana laccase (TRANSPARENT TESTA10) was shown to be involved in the polymerization of flavonoids in the seed coat [41]. The role of multicopper oxidases is more puzzling. The A. thaliana SKU5 (SKEWED5) gene was shown to be involved in root directional growth [42]. Mutants impaired in SKS11 and SKS12 (SKU SIMILAR11 and 12) showed alteration in pollen tube integrity, growth and guidance as well as some alteration in polysaccharide composition [43]. No enzymatic activity has been demonstrated yet for the encoded proteins. Finally, the role of BBE proteins start to be understood thanks to the characterization of the enzymatic activity of the A. thaliana OGOX1-4 (oligogalacturonide OXIDASE 1-4) proteins [44]. They oxidize OGs which are less hydrolysable by fungal PGs and have reduced ability to activate immune response. However, no specific role has yet been demonstrated during plant development.
Several protein families related to lipid metabolism could be identified in most cell wall proteomes. Several roles have been proposed for non-specific lipid transfer proteins (LTPs) [45]. They have been assumed to contribute to the transfer of lipids which are hydrophobic molecules through the hydrophilic cell wall [46]. Indeed, A. thaliana mutants impaired in LTPG2 or in LTPG1 and LTPG2 exhibit an alteration in cuticular wax composition in stems and siliques [47]. LTPG1 and LTPG2 are predicted to be GPI-anchored proteins. LTPs have also been shown to be involved in the adhesion of the cuticular layer on the hydrophilic primary cell wall [48]. Several roles were proposed for GDSL lipases/acylhydrolases [49]. The tomato GDSL1 was shown to be involved in the deposition of cutin in the cuticle of tomato fruits [50]. Indeed, the silencing of GDSL1 leads to the appearance of nanopores in isolated fruit cutins and to a reduction in ester bond cross-links. An A. thaliana mutant impaired in GELP77 exhibits shrunken pollen grains which stick together, suggesting a role of GELP77 in pollen grain wall formation [51]. More recently, GDSL lipases/acylhydrolases were assumed to also be involved in suberin degradation [52].
Among the miscellaneous proteins, dirigent proteins (DIRs) are assumed to be involved in lignan and in lignin biosynthesis. They have no known enzymatic activity, but they would control the regio- and stereoselectivity of bimolecular phenoxy radical coupling [17]. As an example, the A. thaliana AtDIR10 protein was shown to be essential for the establishment of the lignin-based Casparian strips in roots [53]. Several types of enzymatic activities have been associated to germins and germin-like proteins: manganese superoxide dismutase (SOD), oxalate oxidase (OXO) or ADP glucose pyrophosphatase/phosphodiesterase (AGPPase) [54][55]. Thaumatins and thaumatin-like proteins belong to the large pathogenesis-related protein family (PR proteins) and are also called PR-5 [56]. Most of them exhibit an anti-fungal activity and their genes are induced upon biotic stress. They might also have allergenic properties. Extracellular purple acid phosphatases (PAPs) are phosphohydrolases able to cleave Pi from organic Pi-esters that are inaccessible to root cells in soils, for example [18]. The predominant A. thaliana PAPs (AtPAP12 and AtPAP26) were identified in several cell wall proteomes [57][58][59][60] and both proteins were isolated from the culture medium of cell suspensions cultures [61].
Fasciclin arabinogalactan proteins (FLAs) are assumed to be involved in the interactions between the cells and their environment in the same way as mammalian proteins carrying fasciclin domains (FAS1) [62]. Some of them are located at the plasma membrane surface thanks to the presence of a GPI-anchor as experimentally demonstrated for AtFLA4 and AtFLA12 [63][64]. They could also be released in the cell wall after GPI-anchor cleavage. AtFLA4 was assumed to interact with pectin molecules and to contribute to the biomechanical properties of the cell wall [62]. FLAs were also found to be present in the so-called G-layer of tension wood. In particular, mutants impaired in AtFLA11 and AtFLA12 exhibit reduced tensile strength and stiffness [65]. In this case, interactions between FLAs and cellulose microfibrils were suspected. Furthermore, in the functional class comprising signaling molecules, proteins with leucine-rich repeats (LRRs) are found in all cell wall proteomes. Their role is not clear but they could interact with other proteins or with peptides. Such interactions have been reported for the LRR domains of AtLRX2 and AtLRX8 interacting with the rapid alkalinization factor 4 (RALF4) signaling peptide [66].
The DUF 642 (domain of unknown function 642, InterPro domain IPR006946) proteins were initially identified as major proteins in the cell wall proteome of A. thaliana etiolated hypocotyls [58]. The DUF 642 domain is frequently associated with a galactose-binding-like domain (InterPro domain IPR008979). Different roles were proposed, such as a structural role as lectin-like proteins interacting with cell wall polysaccharides [67] or a role in the regulation of PME activity [68].
2. What about the Non-Canonical Proteins Identified in Cell Wall Proteomes?
All the published proteomes characterized from purified cell walls, extracellular fluids or cell suspension culture media contain proteins which are not expected to be secreted. These proteins have now been included in a new version of the plant cell wall proteome database called WallProtDB-2 (https://www.polebio.lrsv.ups-tlse.fr/WallProtDB/) (accessed on 6 april 2022) to allow obtaining an overview of their predicted sub-cellular localization and biological activity. Apart from the 4292 proteins considered to be bona fide CWPs, WallProtDB-2 now contains 6462 proteins presumed to be intracellular and identified in apoplastic fluids or among proteins extracted from purified cell walls. These proteins are assumed to be non-canonical CWPs. To the knowledge, this is the first time that this information has been collected.
In the following, 12 cell wall proteomes have been taken into account. Altogether, they comprise 6425 presumed contaminants proteins. The B. oleracea and the P. patens proteomes have been excluded because the former is a xylem sap proteome and the latter is very small one.
Using TargetP (https://services.healthtech.dtu.dk/service.php?TargetP-2.0) and Predotar (https://urgi.versailles.inrae.fr/predotar/), the proteins presumed to be contaminants were predicted to be targeted to chloroplasts (between 19.9 and 24.8%), mitochondria (between 11.3 and 13.3%), the secretory pathway (between 6.9 and 7.8%) and other cell compartments (between 54.0 and 61.7%). In the case of proteins predicted to be targeted to the secretory pathway, some of them have ER retention signals, or multiple transmembrane-domains such as transporters (7.8% as predicted by TMHMM, https://services.healthtech.dtu.dk/service.php?TMHMM-2.0).
A very high number of domains could be predicted in the proteins presumed to be contaminant: 1575 Pfam (https://xfam.org/) (accessed on 6 April 2022) and 3024 IPR (https://www.ebi.ac.uk/interpro/) (accessed on 6 April 2022) domains. This result shows the huge diversity of these proteins. One third of the Pfam domains (560) were only present in one protein whereas 6 domains were shared by more than 50 proteins . Similar results were observed for IPR domains with 938 domains (about one third) only present in one protein and 36 domains present in more than 50 proteins. The number of proteins sharing a given domain increases with the number of presumed contaminants in a given cell wall proteome.
The frequent identification of certain proteins in cell wall proteomes may have different explanations: (i) they could exhibit specific features allowing them to strongly interact with cell wall components during the purification of cell walls, for example, the histones (61 entries in 7 plant species, PF00125, IPR007125), which are basic proteins like most CWPs [1]; (ii) they could be very abundant proteins such as ribosomal proteins (altogether 578 entries in 8 plant species); or (iii) secreted through alternative secretory pathways. For some protein families, there is no clear hypothesis regarding their presence in many cell wall proteomes: for example, thioredoxin (for example, PF00085 with 117 occurrences in 12 plant species), heat-shock proteins (for example, PF00012 with 67 proteins in 12 plant species), glyceraldehyde 3-phosphate dehydrogenase (PF02800 and PF00044 with 46 and 45 proteins in 11 and 10 plant species, respectively), lactate/malate dehydrogenase (PF02866 and PF00056 with 49 proteins in 10 plant species) and cyclophilin type peptidyl-prolyl cis-trans isomerase (42 proteins in 9 plant species). Finally, these proteins could be moonlighting ones, being present in different cell compartments and having different functions in each of them [69]. As an example, two non-specific lipid transfer proteins of A. thaliana, AtLTP2 and AtLTP4, have been localized in both the cell wall and chloroplasts [48][63].
As mentioned above, UPS pathways have been described in bacteria and mammals. In plants, the best documented example of the presence of leaderless proteins in the apoplast is probably that of the leaderless jacalin-related lectin of Helianthus annuus (Helja): it has been identified in extracellular fluids [70], and in extracellular vesicles [71], and it has been immunolocalized in the extracellular matrix [70]. Another example is that of the cytoplasmic mannitol dehydrogenase which has been immunolocalized in cell walls upon a salicylic treatment [72].
As for mammalian cells, four main UPS pathways have been proposed in plants [73]: a direct ER to plasma membrane traffic, plasma membrane transporter channels, secretory lysosomes, and multivesicular bodies (MVBs) leading to exosome secretion. Besides, exocyst positive organelles (EXPOs) with a double membrane have been characterized in A. thaliana and Nicotiana tabacum cells [74]. Exocysts are proteins mediating the fusion between post-Golgi vesicles and the plasma membrane, thus allowing the release of proteins in the extracellular space. All these pathways are resistant to brefeldin A which disrupts the ER-Golgi vesicular traffic. However, it must be stressed that additional efforts has to be done to better define what is presently called extracellular vesicles (EVs) and to identify specific markers to allow comparing different ones [75].
Recent it has been devoted to EVs in A. thaliana and H. annuus upon pathogen infection or in response to salicylic acid treatment [71][76], and in Nicotiana benthamiana upon viral infection [77]. These vesicles contain proteins involved in plant defense reactions, in membrane trafficking; among which are proteins with or without predicted signal peptides. They have also been shown to deliver small RNAs to fungal pathogens [78] and viral components in the cell wall [77]. Whether these EVs are EXPOs and whether plants produce different kinds of EVs remain to be determined [75][79].
Unfortunately, no bioinformatic program similar to SecretomeP has yet been designed for plant proteins. In this bioinformatic program, it is assumed that proteins present in extracellular spaces share common features whatever the route of secretion [80]. Such a tool would be useful to help sort the proteins devoid of a predictable signal peptide and focusing experiment them to demonstrate their actual presence in apoplastic fluids or in cell walls.
3. Conclusions
The set of conserved CWPs could be involved in housekeeeping functions and/or quick answers to environmental stresses. Studying additional cell wall proteomes would conribute giving a more precise description of this core proteome and scale it down to the organ level. Regarding the non-canonical proteins, experiment will be required to check their actual presence in cell walls and to provide precise descriptions of plant alternative secretion pathways. Among the next challenges for plant cell wall proteomics are the increase of their coverage including the identification of peptides, knowledge of protein half-lives, and a refined description of CWP post-translational modifications. Besides, the integration of transcriptomics and proteomics data will be critical to understand the regulation of the expression of the genes encoding CWPs.
References
- Jamet, E.; Albenne, C.; Boudart, G.; Irshad, M.; Canut, H.; Pont-Lezica, R. Recent advances in plant cell wall proteomics. Proteomics 2008, 8, 893–908.
- Cosgrove, D. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172.
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495.
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 2019, 26, 101247.
- Van der Hoorn, R. Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223.
- Schaller, A.; Stintzi, A.; Rivas, S.; Serrano, I.; Chichkova, N.V.; Vartapetian, A.B.; Martınez, D.; Guiamet, J.J.; Sueldo, D.J.; van der Hoorn, R.A.L.; et al. From structure to function—A family portrait of plant subtilases. New Phytol. 2018, 218, 901–915.
- Edstam, M.; Viitanen, L.; Salminen, T.; Edqvist, J. Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 2011, 4, 947–964.
- Dong, X.; Yi, H.; Han, C.; Nou, I.; Hur, Y. GDSL esterase/lipase genes in Brassica rapa L.: Genome-wide identification and expression analysis. Mol. Genet. Genom. 2016, 291, 531–542.
- Hayashi, S.; Ishii, T.; Matsunaga, T.; Tominaga, R.; Kuromori, T.; Wada, T.; Shinozaki, K.; Hirayama, T. The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol. 2008, 49, 1522–1535.
- Zhou, K. Glycosylphosphatidylinositol-anchored proteins in Arabidopsis and one of their common roles in signaling transduction. Front. Plant Sci. 2019, 10, 1022.
- Seifert, G.J.; Roberts, K. The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 2007, 58, 137–161.
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell 2015, 27, 2095–2118.
- Bellande, K.; Bono, J.-J.; Savelli, B.; Jamet, E.; Canut, H. Plant lectins and lectin receptor-like kinases: How do they sense the outside? Int. J. Mol. Sci. 2017, 18, 1164.
- Okuda, S. Molecular mechanisms of plant peptide binding to receptors. Peptides 2021, 144, 170614.
- Reymond, P. Receptor kinases in plant responses to herbivory. Curr. Opin. Biotechnol. 2021, 70, 143–150.
- Chen, Y.; Ye, D.; Held, M.A.; Cannon, M.C.; Tui, R.; Saha, P.; Frye, A.N.; Mort, A.J.; Kieliszewski, M.J. Identification of the abundant hydroxyproline-rich glycoproteins in the root walls of wild-type Arabidopsis, an ext3 mutant line, and its phenotypic revertant. Plants 2015, 4, 85–111.
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301.
- Dissanayaka, D.; Ghahremani, M.; Siebers, M.; Wasaki, J.; Plaxton, W. Recent insights into the metabolic adaptations of phosphorus-deprived plants. J. Exp. Bot. 2021, 72, 199–223.
- Sousa, A.O.; Assis, E.T.; Pirovani, C.P.; Alvim, F.C.; Costa, M.G. Phosphate-induced-1 gene from Eucalyptus (EgPHI-1) enhances osmotic stress tolerance in transgenic tobacco. Genet. Mol. Res. 2014, 13, 1579–1588.
- Wang, T.; Chen, X.; Zhu, F.; Li, H.; Li, L.; Yang, Q.; Chi, X.; Yu, S.; Liang, X. Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PLoS ONE 2013, 8, e61722.
- Fry, S.C. Primary cell wall metabolism: Tracking the careers of wall polymers in living plant cells. New Phytol. 2004, 161, 641–675.
- Shinohara, N.; Sunagawa, N.; Tamura, S.; Yokoyama, R.; Ueda, M.; Igarashi, K.; Nishitani, K. The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cello-oligosaccharide. Sci. Rep. 2017, 7, 46099.
- Strohmeier, M.; Hrmova, M.; Fischer, M.; Harvey, A.; Fincher, G.; Pleiss, J. Molecular modeling of family GH16 glycoside hydrolases: Potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the poaceae. Protein Sci. 2009, 13, 3200–3213.
- Liu, Y.; Lu, S.; Zhang, J.; Liu, S.; Lu, Y. A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 2007, 226, 1547–1560.
- Minic, Z.; Jouanin, L. Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol. Biochem. 2006, 44, 435–449.
- Levy, A.; Erlanger, M.; Rosenthal, M.; Epel, B. A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J. 2007, 49, 669–682.
- Rhee, S.; Osborne, E.; Poindexter, P.; Somerville, C. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 2003, 133, 1170–1180.
- Hocq, L.; Pelloux, J.; Lefebvre, V. Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 2017, 22, 20–29.
- Guénin, S.; Mareck, A.; Rayon, C.; Lamour, R.; Assoumou Ndong, Y.; Domon, J.; Sénéchal, F.; Fournet, F.; Jamet, E.; Canut, H.; et al. Identification of pectin methylesterase 3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis thaliana. New Phytol. 2011, 192, 114–126.
- Minic, Z. Physiological roles of plant glycoside hydrolases. Planta 2008, 227, 723–740.
- Van Hengel, A.; Tadesse, Z.; Immerzeel, P.; Schols, H.; van Kammen, A.; De Vries, S. N-acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol. 2001, 117, 1880–1890.
- Orlando, M.; Buchholz, P.; Lotti, M.; Pleiss, J. The GH19 Engineering Database: Sequence diversity, substrate scope, and evolution in glycoside hydrolase family 19. PLoS ONE 2021, 16, e0256817.
- Kesari, P.; Narhari Patil, D.; Kumar, P.; Tomar, S.; Kumar Sharma, A.; Kumar, P. Structural and functional evolution of chitinase-like proteins from plants. Proteomics 2015, 15, 1693–1705.
- Wan, H.; Wu, L.; Yang, Y.; Zhou, G.; Ruan, Y. Evolution of sucrose metabolism: The dichotomy of invertases and beyond. Trends Plant Sci. 2018, 23, 163–177.
- Proels, R.; Hückelhoven, R. Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol. Plant Pathol. 2014, 15, 858–864.
- Stührwohldt, N.; Ehinger, A.; Thellmann, K.; Schaller, A. Processing and formation of bioactive CLE40 peptide are controlled by posttranslational proline hydroxylation. Plant Physiol Biochem 2020, 184, 1573–1584.
- Von Groll, U.; Berger, D.; Altmann, T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 2002, 14, 1527–1539.
- Xia, Y.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988.
- Francoz, E.; Ranocha, P.; Nguyen-Kim, H.; Jamet, E.; Burlat, V.; Dunand, C. Roles of cell wall peroxidases in plant development. Phytochemistry 2015, 112, 15–21.
- Wang, Y.; Bouchabke-Coussa, O.; Lebris, P.; Antelme, S.; Soulhat, C.; Gineau, E.; Dalmais, M.; Bendahmane, A.; Morin, H.; Mouille, G.; et al. LACCASE5 is required for lignification of the Brachypodium distachyon culm. Plant Physiol. 2015, 168, 192–204.
- Pourcel, L.; Routaboul, J.M.; Kerhoas, L.; Caboche, M.; Lepiniec, L.; Debeaujon, I. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 2005, 17, 2966–2980.
- Sedbrook, J.C.; Carroll, K.L.; Hung, K.F.; Masson, P.H.; Somerville, C.R. The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 2002, 14, 1635–1648.
- Duan, Y.; Wang, L.; Li, X.; Wang, W.; Wang, J.; Liu, X.; Zhong, Y.; Cao, N.; Tong, M.; Ge, W.; et al. Arabidopsis SKU5 Similar 11 and 12 play crucial roles in pollen tube integrity, growth and guidance. Plant J. 2021, 109, 598–614.
- Benedetti, M.; Verrascina, I.; Pontiggia, D.; Locci, F.; Mattei, B.; De Lorenzo, G.; Cervone, F. Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J. 2018, 94, 260–273.
- Jacq, A.; Burlat, V.; Jamet, E. Plant cell wall proteomics as a strategy to reveal candidate proteins involved in extracellular lipid metabolism. Curr. Protein Pept. Sci. 2018, 19, 190–199.
- DeBono, A.; Yeats, T.; Rose, J.; Bird, D.; Reinhard Jetter, R.; Kunst, L.; Samuels, L. Arabidopsis LTPG Is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 2009, 21, 1230–1238.
- Kim, H.; Lee, S.; Kim, H.; Min, M.; Hwang, I.; Suh, M. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell. Physiol. 2012, 53, 1391–1403.
- Jacq, A.; Pernot, C.; Martinez, Y.; Domergue, F.; Payré, B.; Jamet, E.; Burlat, V.; Pacquit, V. The Arabidopsis Lipid Transfer Protein 2 (AtLTP2) is involved in cuticle-cell wall interface integrity and in etiolated hypocotyl permeability. Front. Plant Sci. 2017, 8, 263.
- Ding, L.; Li, M.; Wang, W.; Cao, J.; Wang, Z.; Zhui, K.; Yang, Y.; Li, Y.; Tan, X. Advances in plant GDSL lipases: From sequences to functional mechanisms. Acta Physiol. Plant. 2019, 41, 151.
- Girard, A.L.; Mounet, F.; Lemaire-Chamley, M.; Gaillard, C.; Elmorjani, K.; Vivancos, J.; Runavot, J.L.; Quemener, B.; Petit, J.; Germain, V.; et al. Tomato GDSL 1 is required for cutin deposition in the fruit cuticle. Plant Cell 2012, 24, 3119–3134.
- Tsugama, D.; Fugino, K.; Liu, S.; Takano, T. GDSL-type esterase/lipase gene, GELP77, is necessary for pollen dissociation and fertility in Arabidopsis. Biochem. Biophys. Res. Commun. 2020, 526, 1036–1041.
- Ursache, R.; De Jesus Vieira Teixeira, C.; Dénervaud Tendon, V.; Gully, K.; De Bellis, D.; Schmid-Siegert, E.; Andersen, T.; Shekhar, V.; Calderon, S.; Pradervand, S.; et al. GDSL-domain proteins have key roles in suberin polymerization and degradation. Nat. Plants 2021, 7, 353–364.
- Hosmani, P.; Kamiya, T.; Danku, J.; Naseer, S.; Geldner, N.; Guerinot, M.; Salt, D. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. USA 2013, 110, 14498–14503.
- Dunnwell, J.; Gibbings, J.; Mahmood, T.; Naqvi, S. Germin and germin-like proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 2008, 27, 342–375.
- Barman, A.; Banerjee, J. Versatility of germin-like proteins in their sequences, expressions, and functions. Funct. Integr. Genom. 2015, 15, 533–548.
- De Jesús-Pires, C.; Costa Ferreira-Neto, J.; Bezerra-Neto, J.; Kido, E.; de Oliveira Silva, R.; Pandolfi, V.; Wanderley-Nogueira, A.; Binneck, E.; da Costa, A.; Pio-Ribeiro, G.; et al. Plant thaumatin-like proteins: Function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 2020, 21, 36–51.
- Duruflé, H.; Ranocha, P.; Balliau, T.; Dunand, C.; Jamet, E. Transcriptomic and cell wall proteomic datasets of rosettes and floral stems from five Arabidopsis thaliana ecotypes grown at optimal or sub-optimal temperature. Data Brief 2019, 27, 104581.
- Irshad, M.; Canut, H.; Borderies, G.; Pont-Lezica, R.; Jamet, E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 2008, 8, 94.
- Nguyen-Kim, H.; San Clemente, H.; Balliau, T.; Zivy, M.; Dunand, C.; Albenne, C.; Jamet, E. Arabidopsis thaliana root cell wall proteomics: Increasing the proteome coverage using a combinatorial peptide ligand library and description of unexpected Hyp in peroxidase amino acid sequences. Proteomics 2016, 16, 491–503.
- Xu, S.; Medzihradszky, K.; Wang, Z.; Burlingame, A.; Chalkley, R. N-Glycopeptide profiling in Arabidopsis inflorescence. Mol. Cell. Proteom. 2016, 15, 2048–2054.
- Tran, H.T.; Qian, W.; Hurley, B.A.; She, Y.M.; Wang, D.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP12 and AtPAP26: The predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ. 2010, 33, 1789–1803.
- Seifert, G. Fascinating fasciclins: A surprisingly widespread family of proteins that mediate interactions between the cell exterior and the cell surface. Int. J. Mol. Sci. 2018, 19, 1628.
- Pinski, A.; Roujol, D.; Pouzet, C.; Bordes, L.; San Clemente, H.; Hoffmann, L.; Jamet, E. Comparison of mass spectrometry data and bioinformatics predictions to assess the bona fide localization of proteins identified in cell wall proteomics studies. Plant Sci. 2021, 310, 110979.
- Griffiths, J.; Crepeau, M.; Ralet, M.; Seifert, G.; North, H. Dissecting seed mucilage adherence mediated by FEI and SOS5. Front. Plant Sci. 2016, 7, 1073.
- MacMillan, C.; Taylor, L.; Bi, Y.; Southerton, S.; Evans, R.; Spokevicius, A. The fasciclin-like arabinogalactan protein family of Eucalyptus grandis contains members that impact wood biology and biomechanics. New Phytol. 2015, 206, 1314–1327.
- Moussu, S.; Broyart, C.; Santos-Fernandez, G.; Augustin, S.; Wehrle, S.; Grossniklaus, U.; Santiago, J. Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc. Natl. Acad. Sci. USA 2020, 117, 7494–7503.
- Vázquez-Lobo, A.; Roujol, D.; Zuñiga-Sánchez, E.; Albenne, C.; Piñero, D.; de Buen, A.G.; Jamet, E. The highly conserved spermatophyte cell wall DUF642 protein family: Phylogeny and first evidence of interaction with cell wall polysaccharides in vitro. Mol. Phylogenet. Evol. 2012, 63, 510–520.
- Cruz-Valderrama, J.; Gómez-Maqueo, X.; Salazar-Iribe, A.; Zúñiga-Sánchez, E.; Hernández-Barrera, A.; Quezada-Rodríguez, E.; Gamboa-deBuen, A. Overview of the role of cell wall DUF642 proteins in plant development. Int. J. Mol. Sci. 2019, 20, 3333.
- Jeffery, C.J. Mass spectrometry and the search for moonlighting proteins. Mass Spectrom. Rev. 2005, 24, 772–782.
- Pinedo, M.; Regente, M.; Elizalde, M.; Quiroga, I.; Pagnussat, L.A.; Jorrin-Novo, J.; Maldonado, A.; de la Canal, L. Extracellular sunflower proteins: Evidence on non-classical secretion of a jacalin-related lectin. Protein Pept. Lett. 2012, 19, 270–276.
- Regente, M.; Pinedo, M.; San Clemente, H.; Balliau, T.; Jamet, E.; de la Canal, L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot. 2017, 68, 5485–5495.
- Cheng, F.; Zamski, E.; Guo, W.; Pharr, D.; Williamson, J. Salicylic acid stimulates secretion of the normally symplastic enzyme mannitol dehydrogenase: A possible defense against mannitol-secreting fungal pathogens. Planta 2009, 230, 1093–1103.
- Rose, J.K.C.; Lee, S.-J. Straying off the highway: Trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol. 2010, 153, 433–436.
- Wang, J.; Ding, Y.; Wang, J.; Hillmer, S.; Miao, Y.; Lo, S.; Wang, X.; Robinson, D.; Jiang, L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 2010, 22, 4009–4030.
- Pinedo, M.; de la Canal, L.; de Marcos Lousa, C. A call ofr rigor and standardization in plant extracellular vesicle research. J. Extracell Vesicles 2021, 10, e12048.
- Rutter, B.; Innes, R. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 2017, 173, 728–741.
- Movahed, N.; Garcia Cabanillas, D.; Wan, J.; Vali, H.; Laliberté, J.F.; Zheng, H. Turnip Mosaic Virus components are released into the extracellular space by vesicles in infected leaves. Plant Physiol. 2021, 180, 1375–1388.
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.; Palmquist, J.; Huang, S.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129.
- Boevink, P. Exchanging missives and missiles: The roles of extracellular vesicles in plant–pathogen interactions. J. Exp. Bot. 2018, 68, 5411–5414.
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; von Heijne, G.; Brunak, S. Feature based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
960
Revisions:
2 times
(View History)
Update Date:
21 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




