Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Athena Davri | -- | 1812 | 2022-04-14 15:41:46 | | | |
| 2 | Lindsay Dong | -9 word(s) | 1803 | 2022-04-15 07:32:56 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Davri, A.; Giannakeas, N.; Birbas, E.; Kanavos, T.; Ntritsos, G.; Tzallas, A.; Batistatou, A. Deep Learning on Images for Colorectal Cancer Diagnosis. Encyclopedia. Available online: https://encyclopedia.pub/entry/21779 (accessed on 07 February 2026).
Davri A, Giannakeas N, Birbas E, Kanavos T, Ntritsos G, Tzallas A, et al. Deep Learning on Images for Colorectal Cancer Diagnosis. Encyclopedia. Available at: https://encyclopedia.pub/entry/21779. Accessed February 07, 2026.
Davri, Athena, Nikolaos Giannakeas, Effrosyni Birbas, Theofilos Kanavos, Georgios Ntritsos, Alexandros Tzallas, Anna Batistatou. "Deep Learning on Images for Colorectal Cancer Diagnosis" Encyclopedia, https://encyclopedia.pub/entry/21779 (accessed February 07, 2026).
Davri, A., Giannakeas, N., Birbas, E., Kanavos, T., Ntritsos, G., Tzallas, A., & Batistatou, A. (2022, April 14). Deep Learning on Images for Colorectal Cancer Diagnosis. In Encyclopedia. https://encyclopedia.pub/entry/21779
Davri, Athena, et al. "Deep Learning on Images for Colorectal Cancer Diagnosis." Encyclopedia. Web. 14 April, 2022.
Copy Citation
Colorectal cancer (CRC) is the second most common cancer in women and the third most common in men, with an increasing incidence. Pathology diagnosis complemented with prognostic and predictive biomarker information is the first step for personalized treatment. Artificial Intelligence (AI) has made significant progress in the medical field, showing potential for clinical applications.
colorectal cancer
CRC
microscopy images
deep learning
DL
convolutional neural networks
CNN
artificial intelligence
1. Introduction
Colorectal cancer (CRC) is one of the most common types of gastrointestinal cancer, the second most common cancer in women and the third in men [1]. Despite existing variations, such as geographical distribution, age and gender differences, the CRC incidence, overall, is estimated to increase by 80% in the year 2035, worldwide [2]. This rising incidence of CRC is mainly due to changes in lifestyle, particularly dietary patterns [3]. Most CRCs are sporadic (70–80%), while approximately one third have a hereditary component [4]. Within the term CRC, a wide range of carcinoma subtypes is included, characterized by different morphological features and molecular alterations.
The cornerstone of CRC diagnosis is the pathologic examination (biopsy or surgical excision) [5]. With the advent of screening methods, many precursor lesions are also detected and biopsied. Consequently, a wide range of pre-malignant lesions have been identified, and occasionally, a differential diagnosis between pre-malignant and malignant lesions is quite challenging [6]. The histopathological examination of the tissue remains the “gold standard” for diagnosis, with the first step being the optimal preparation of the histological section, stained with Hematoxylin and Eosin (H&E) [7]. Further examination with special in situ methods, such as immunohistochemistry (IHC) and in situ hybridization (ISH), and other molecular techniques follows [8]. There are published guidelines for pre-analytical, analytical and post-analytical procedures in a pathology laboratory [9]. As expected, due to the high incidence of CRC, the diagnostic load in a routine pathology laboratory is very heavy and the introduction of an ever-growing list of morpho-molecular features to be examined and noted has made the diagnosis a time-consuming process [10]. All these factors, in combination with the shortage of pathologists worldwide, have led to delays in diagnosis, with consequences to the optimal healthcare of the patient.
It has been shown that pathologists make a diagnosis based mainly on image-based pattern recognition [6]. With this strategy, architectural and cellular characteristics conform to already known features of a disease [11]. In several instances, an accurate diagnosis or estimation of prognostic and predictive factors is subject to personal interpretations, leading to inter- and intra-observer variability [12][13]. In a continuous effort to improve the accuracy of the pathology diagnosis, combined with the timely delivery of all vital information for optimal patient treatment, the new and breakthrough technologies can be of great value.
The recent World Health Organization (WHO) classification for malignant epithelial tumors of the colorectum includes four main categories: adenocarcinoma (ADC) not otherwise specified (NOS), neuroendocrine tumor NOS, neuroendocrine carcinoma NOS and mixed neuroendocrine-non-neuroendocrine neoplasm (MiMEN) [14]. Of these, colorectal ADC is the most common (90%) and, by definition, it shows glandular and mucinous differentiation. Colorectal ADC has several histopathological subtypes, with specific morphologic, clinical, and molecular characteristics, i.e., serrated ADC, adenoma-like ADC, micropapillary ADC, mucinous ADC, poorly cohesive carcinoma, signet-ring cell carcinoma, medullary ADC, adenosquamous carcinoma, carcinoma undifferentiated NOS and carcinoma with sarcomatoid component.
The diagnosis of CRC is only the first step for a complete pathology report. According to best-practice guidelines, the specific histologic subtype, the histologic grade, the TNM staging system, the lymphovascular and perineural invasion, and the tumor budding should be reported [9][14].
Histopathology image generations start with the standard procedure of tissue preparation. Biopsy or surgical specimens (representative sections) are formalin-fixed and paraffin-embedded. Then, the 4μm tissue sections are prepared and stained with H&E dye [15]. The images are extracted after a scanning procedure. Several scanning systems can be used to digitize the whole slide [16], such as the Hamamatsu NanoZoomer series, the Omnyx scanner, the Zeiss scanners, the Pannoramic 250 Flash II, and the Leica Biosystems Aperio systems [17]. Most of the above scanners provide two optical magnifications, 20× and 40×, however, the user can also digitally undersample the image in different magnifications. A scanner needs several minutes for the scanning of the whole slide, while most of the system can deal with tens or hundreds of slides that are scanned automatically one-by-one. According to the digitalization, each pixel of a Whole Slide Image (WSI) corresponds to a physical area of several decades nm2. For example, in the 40× magnification mode, a Hamamatsu NanoZoomer scanner extracts an image, where the size of each pixel edge corresponds to 227 nm [18]. The latter image digitization provides an appropriate resolution for most of the histological findings, which presents a physical size of microns [19]. In most of the cases, the extracting images are storage either in a compressed JPEG-based format or an uncompressed TIFF format. Figure 1 presents the resolution of a WSI, scanned by a Hamamatsu NanoΖoomer 210.
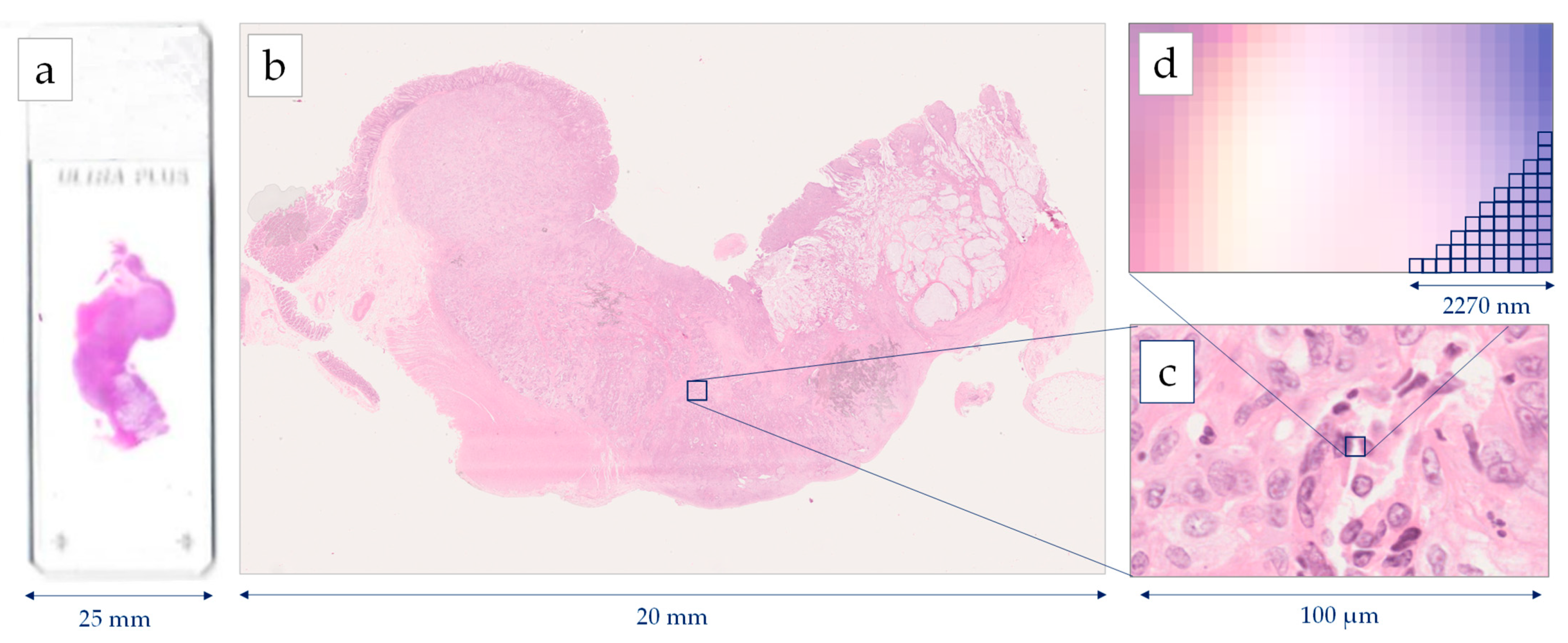
Figure 1. Image generation using a Hamamatsu NanoZoomer whole slide scanner: (a) histological slide 75 mm × 25 mm, (b) Whole Slide Image (WSI), (c) cell level in 40× magnification, (d) pixel level in 40× magnification digitizing images 227 nm per pixel.
Machine learning is a branch of AI which is based on the concept that machines could have access to data and be able to learn on their own. AI has a broader scope and involves machines that are capable of carrying out tasks requiring intelligence. Machine learning techniques focus on the creation of intelligent software using statistical learning methods and require access to data for the learning procedure [20]. A branch of machine learning, which has drawn a lot of attention over the last few years, is DL. DL involves training artificial neural networks (ANNs) with multiple layers of artificial neurons (nodes). Neural networks are inspired from the human physiology of the brain, comprising a simplified artificial model of the human neural network. An ANN is a collection of connected artificial neurons. The simplest ANN architecture is the single layer feed forward neural network. In these types of networks, the information moves in one direction only, from the inputs’ nodes to the hidden layer nodes and then to the output nodes. The success and wide acceptance of ANNs relies on their capability to solve complex mathematical problems, nonlinear or stochastic, by using very simple computational operations. In contrast to a conventional algorithm, which needs complex mathematical and algorithmic operations and could only apply to one problem, an ANN is computationally and algorithmically very simple and its structure allows it to be applied in a wide range of problems [21].
2. Deep Learning on Images for Colorectal Cancer Diagnosis
A pathology diagnosis focuses on the macroscopic and microscopic examination of human tissues, with the light microscope being the valuable tool for almost two centuries [11]. A meticulous microscopic examination of tissue biopsies is the cornerstone of diagnosis and is a time-consuming procedure. An accurate diagnosis is only the first step for patient treatment. It needs to be complimented with information about grade, stage, and other prognostic and predictive factors [4]. Pathologists’ interpretations of tissue lesions become data, guiding decisions for patients’ management. A meaningful interpretation is the ultimate challenge. In certain fields, inter- and intra-observer variability are not uncommon [12][13]. In such cases, the interpretation of the visual image can be assisted by objective outputs. Many data have been published over the last 5 years exploring the possibility of moving on to computer-aided diagnosis and the measurement of prognostic and predictive markers for optimal personalized medicine [22][23]. Furthermore, the implementation of AI is now on the horizon. In the last 5 years, extensive research has been conducted to implement AI-based models for the diagnosis of multiple cancer types and, in particular, CRC [24][25][26]. The important aspects in a CRC diagnosis, such as histological type, grade, stromal reaction, immunohistochemical and molecular features have been addressed using breakthrough technologies.
The traditional pathology methods are accompanied by great advantages [27]. The analytical procedures in pathology laboratories are cost-effective and, during recent years, have become automated, eliminating the time and errors of procedures, while maintaining high levels of sensitivity and specificity of techniques, such as IHC [26]. Despite the widespread availability, challenges and limitations of traditional pathology methods remain, such as the differences between laboratories’ protocols and techniques, as well as the subjective interpretation between pathologists, resulting in inconsistency in diagnoses [12][13]. Novel imaging systems and WSI scanners promise to upgrade traditional pathology, preserving the code and ethics of practice [26]. The potential of DL algorithms is expanding all over the fields in histopathology. In clinical practice, such algorithms could provide valuable information about the tumor microenvironment quantitative analysis of histological features [28]. Better patient stratification for targeted therapies could be approached by DL-based models predicting mutations, such as MSI status [29][30][31]. More than ever, AI could be of great importance for a pathologist in daily clinical practice. AI is consistently supported by extensive research, which is followed by good performance metrics and potential. Several studies have shown that many DL-based models’ predictions did not differ in terms of statistical significance when compared to pathologists’ predictions [32][33]. Thus, DL algorithms could provide valuable results for diagnoses in clinical practice, especially when inconsistencies occur. The available scanned histological images can be reviewed and examined by the collaboration of pathologists simultaneously, from different locations [34][35]. For an efficient fully digital workflow, however, the development of technology infrastructure, including computers, scanners, workstations and medical displays is necessary.
The application of DL methods in the diagnosis of CRC over the last 5-years seems to be evolving rapidly, faster than other fields of histopathology. However, it seems that there is an expected gradual evolution, starting from the simple techniques of CNNs, then employing transfer learning to the networks, and finally attempting to develop new architectures, focusing on the requirements of the medical question. Additionally, in the last two years, alternative deep learning techniques such as GANs have started to be used. The contribution of such methods will be significant, since DL requires a sufficient size of the training set to perform well and provide generalization. Large data sets may not always be available from the annotations of pathologists and, therefore, need to be enriched with a simulated training set.
It is expected that CNN’s application directly in histopathological images will present a better performance compared to traditional techniques. CNNs are advantageous over traditional image processing techniques due to the training procedure, while they are also more robust than the traditional AI techniques because they automatically extract features from the image.
3. Conclusions
When dealing with human disease, particularly cancer, AI is promising to deliver valuable guidance. Specifically for CRC, it appears that the recent exponentially growing relevant research will soon transform the field of tissue-based diagnosis. Preliminary results demonstrate that AI-based models are further applied in clinical cancer research, including CRC, breast and lung cancer. However, to overcome several limitations, larger numbers of datasets, quality image annotations, as well as external validation cohorts are required to establish the diagnostic accuracy of DL models in clinical practice.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Douaiher, J.; Ravipati, A.; Grams, B.; Chowdhury, S.; Alatise, O.; Are, C. Colorectal Cancer-Global Burden, Trends, and Geographical Variations. J. Surg. Oncol. 2017, 115, 619–630.
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025.
- Müller, M.F.; Ibrahim, A.E.K.; Arends, M.J. Molecular Pathological Classification of Colorectal Cancer. Virchows Arch. 2016, 469, 125–134.
- Marzouk, O.; Schofield, J. Review of Histopathological and Molecular Prognostic Features in Colorectal Cancer. Cancers 2011, 3, 2767–2810.
- Jass, J.R. Classification of Colorectal Cancer Based on Correlation of Clinical, Morphological and Molecular Features. Histopathology 2007, 50, 113–130.
- Sideris, M.; Papagrigoriadis, S. Molecular Biomarkers and Classification Models in the Evaluation of the Prognosis of Colorectal Cancer. Anticancer Res. 2014, 34, 2061–2068.
- Pallag, A.; Roşca, E.; Ţiţ, D.M.; Muţiu, G.; Bungău, S.G.; Pop, O.L. Monitoring the Effects of Treatment in Colon Cancer Cells Using Immunohistochemical and Histoenzymatic Techniques. Rom. J. Morphol. Embryol. 2015, 56, 1103–1109.
- Vogel, J.D.; Eskicioglu, C.; Weiser, M.R.; Feingold, D.L.; Steele, S.R. The American Society of Colon and Rectal Surgeons Clinical Practice Guidelines for the Treatment of Colon Cancer. Dis. Colon Rectum 2017, 60, 999–1017.
- Kelly, M.; Soles, R.; Garcia, E.; Kundu, I. Job Stress, Burnout, Work-Life Balance, Well-Being, and Job Satisfaction among Pathology Residents and Fellows. Am. J. Clin. Pathol. 2020, 153, 449–469.
- Pena, G.P.; Andrade-Filho, J.S. How Does a Pathologist Make a Diagnosis? Arch. Pathol. Lab. Med. 2009, 133, 124–132.
- van Putten, P.G.; Hol, L.; van Dekken, H.; Han van Krieken, J.; van Ballegooijen, M.; Kuipers, E.J.; van Leerdam, M.E. Inter-Observer Variation in the Histological Diagnosis of Polyps in Colorectal Cancer Screening. Histopathology 2011, 58, 974–981.
- Smits, L.J.H.; Vink-Börger, E.; Lijnschoten, G.; Focke-Snieders, I.; Post, R.S.; Tuynman, J.B.; Grieken, N.C.T.; Nagtegaal, I.D. Diagnostic Variability in the Histopathological Assessment of Advanced Colorectal Adenomas and Early Colorectal Cancer in a Screening Population. Histopathology 2022, 80, 790–798.
- WHO Classification of Tumours Editorial Board. WHO Classification of Tumors: Digestive System Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2019; ISBN 978-92-832-4499-8.
- Arjmand, A.; Tsipouras, M.G.; Tzallas, A.T.; Forlano, R.; Manousou, P.; Giannakeas, N. Quantification of Liver Fibrosis—A Comparative Study. Appl. Sci. 2020, 10, 447.
- Aeffner, F.; Zarella, M.; Buchbinder, N.; Bui, M.; Goodman, M.; Hartman, D.; Lujan, G.; Molani, M.; Parwani, A.; Lillard, K.; et al. Introduction to Digital Image Analysis in Whole-Slide Imaging: A White Paper from the Digital Pathology Association. J. Pathol. Inform. 2019, 10, 9.
- Patel, A.; Balis, U.G.J.; Cheng, J.; Li, Z.; Lujan, G.; McClintock, D.S.; Pantanowitz, L.; Parwani, A. Contemporary Whole Slide Imaging Devices and Their Applications within the Modern Pathology Department: A Selected Hardware Review. J. Pathol. Inform. 2021, 12, 50.
- Jirik, M.; Gruber, I.; Moulisova, V.; Schindler, C.; Cervenkova, L.; Palek, R.; Rosendorf, J.; Arlt, J.; Bolek, L.; Dejmek, J.; et al. Semantic Segmentation of Intralobular and Extralobular Tissue from Liver Scaffold H&E Images. Sensors 2020, 20, 7063.
- Shapcott, M.; Hewitt, K.J.; Rajpoot, N. Deep Learning With Sampling in Colon Cancer Histology. Front. Bioeng. Biotechnol. 2019, 7, 52.
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; Volume 7, pp. 248–255.
- Graupe, D. Principles of Artificial Neural Networks, Advanced Series in Circuits and Systems; World Scientific: London, UK, 2013; Volume 7.
- van der Laak, J.; Litjens, G.; Ciompi, F. Deep Learning in Histopathology: The Path to the Clinic. Nat. Med. 2021, 27, 775–784.
- Wang, Y.; He, X.; Nie, H.; Zhou, J.; Cao, P.; Ou, C. Application of Artificial Intelligence to the Diagnosis and Therapy of Colorectal Cancer. Am. J. Cancer Res. 2020, 10, 3575–3598.
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial Intelligence in Cancer Diagnosis and Prognosis: Opportunities and Challenges. Cancer Lett. 2020, 471, 61–71.
- Thakur, N.; Yoon, H.; Chong, Y. Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review. Cancers 2020, 12, 1884.
- Montezuma, D.; Monteiro, A.; Fraga, J.; Ribeiro, L.; Gonçalves, S.; Tavares, A.; Monteiro, J.; Macedo-Pinto, I. Digital Pathology Implementation in Private Practice: Specific Challenges and Opportunities. Diagnostics 2022, 12, 529.
- Baxi, V.; Edwards, R.; Montalto, M.; Saha, S. Digital Pathology and Artificial Intelligence in Translational Medicine and Clinical Practice. Mod. Pathol. 2022, 35, 23–32.
- Xu, Z.; Li, Y.; Wang, Y.; Zhang, S.; Huang, Y.; Yao, S.; Han, C.; Pan, X.; Shi, Z.; Mao, Y.; et al. A Deep Learning Quantified Stroma-Immune Score to Predict Survival of Patients with Stage II–III Colorectal Cancer. Cancer Cell Int. 2021, 21, 585.
- Jang, H.J.; Lee, A.; Kang, J.; Song, I.H.; Lee, S.H. Prediction of Clinically Actionable Genetic Alterations from Colorectal Cancer Histopathology Images Using Deep Learning. World J. Gastroenterol. 2020, 26, 6207–6223.
- Bilal, M.; Raza, S.E.A.; Azam, A.; Graham, S.; Ilyas, M.; Cree, I.A.; Snead, D.; Minhas, F.; Rajpoot, N.M. Development and Validation of a Weakly Supervised Deep Learning Framework to Predict the Status of Molecular Pathways and Key Mutations in Colorectal Cancer from Routine Histology Images: A Retrospective Study. Lancet Digit. Health 2021, 3, e763–e772.
- Lee, S.H.; Song, I.H.; Jang, H.J. Feasibility of Deep Learning-Based Fully Automated Classification of Microsatellite Instability in Tissue Slides of Colorectal Cancer. Int. J. Cancer 2021, 149, 728–740.
- Wang, K.S.; Yu, G.; Xu, C.; Meng, X.H.; Zhou, J.; Zheng, C.; Deng, Z.; Shang, L.; Liu, R.; Su, S.; et al. Accurate Diagnosis of Colorectal Cancer Based on Histopathology Images Using Artificial Intelligence. BMC Med. 2021, 19, 76.
- Yamashita, R.; Long, J.; Longacre, T.; Peng, L.; Berry, G.; Martin, B.; Higgins, J.; Rubin, D.L.; Shen, J. Deep Learning Model for the Prediction of Microsatellite Instability in Colorectal Cancer: A Diagnostic Study. Lancet Oncol. 2021, 22, 132–141.
- Oliveira, S.P.; Neto, P.C.; Fraga, J.; Montezuma, D.; Monteiro, A.; Monteiro, J.; Ribeiro, L.; Gonçalves, S.; Pinto, I.M.; Cardoso, J.S. CAD Systems for Colorectal Cancer from WSI Are Still Not Ready for Clinical Acceptance. Sci. Rep. 2021, 11, 14358.
- Yoshida, H.; Kiyuna, T. Requirements for Implementation of Artificial Intelligence in the Practice of Gastrointestinal Pathology. World J. Gastroenterol. 2021, 27, 2818–2833.
More
Information
Subjects:
Primary Health Care
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
825
Revisions:
2 times
(View History)
Update Date:
15 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




