
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Masayuki Yamashita | + 2915 word(s) | 2915 | 2022-02-20 11:20:53 | | | |
| 2 | Jessie Wu | Meta information modification | 2915 | 2022-03-11 01:40:49 | | | | |
| 3 | Jessie Wu | Meta information modification | 2915 | 2022-03-11 01:44:05 | | |
Video Upload Options
Hematopoietic stem cells (HSCs) are the only cell population that possesses both a self-renewing capacity and multipotency, and can give rise to all lineages of blood cells throughout an organism’s life. However, the self-renewal capacity of HSCs is not infinite, and cumulative evidence suggests that HSCs alter their function and become less active during organismal aging, leading ultimately to the disruption of hematopoietic homeostasis, such as anemia, perturbed immunity and increased propensity to hematological malignancies. Thus, understanding how HSCs alter their function during aging is a matter of critical importance to prevent or overcome these age-related changes in the blood system. Recent advances in clonal analysis have revealed the functional heterogeneity of murine HSC pools that is established upon development and skewed toward the clonal expansion of functionally poised HSCs during aging. In humans, next-generation sequencing has revealed age-related clonal hematopoiesis that originates from HSC subsets with acquired somatic mutations, and has highlighted it as a significant risk factor for hematological malignancies and cardiovascular diseases.
1. Introduction
Hematopoietic stem cells (HSCs) are blood-forming stem cells that can clonally expand by self-renewing cell division, and that disseminate their clonal progeny by differentiating to all lineages of the blood and immune cells, such as leukocytes, erythrocytes and platelets [1]. Although some exceptions have been identified recently, such as tissue-resident macrophages [2][3] and innate-like B and T lymphocytes [4], HSCs are an important source for the vast majority of the blood and immune cells throughout an organism’s life, during steady-state hematopoiesis and hematopoietic regeneration after injury, thus being attractive targets for preventing or curing hematopoietic disorders. Even after the establishment and sophistication of efficient HSC purification methods, single-cell-based analyses have revealed a remarkable phenotypic and functional heterogeneity in the HSC pool [5]. Of note, recent advances in the clonal tracking methods, referred to as “fate mapping”, provided evidence that HSC heterogeneity can originate from individual clones that emerge during the developmental process [6][7]. Moreover, clonal analyses have revealed an age-related expansion of functionally skewed HSCs, which is associated with an imbalanced production between blood lineages, and reduced clonal diversity in hematopoiesis [6][8][9].
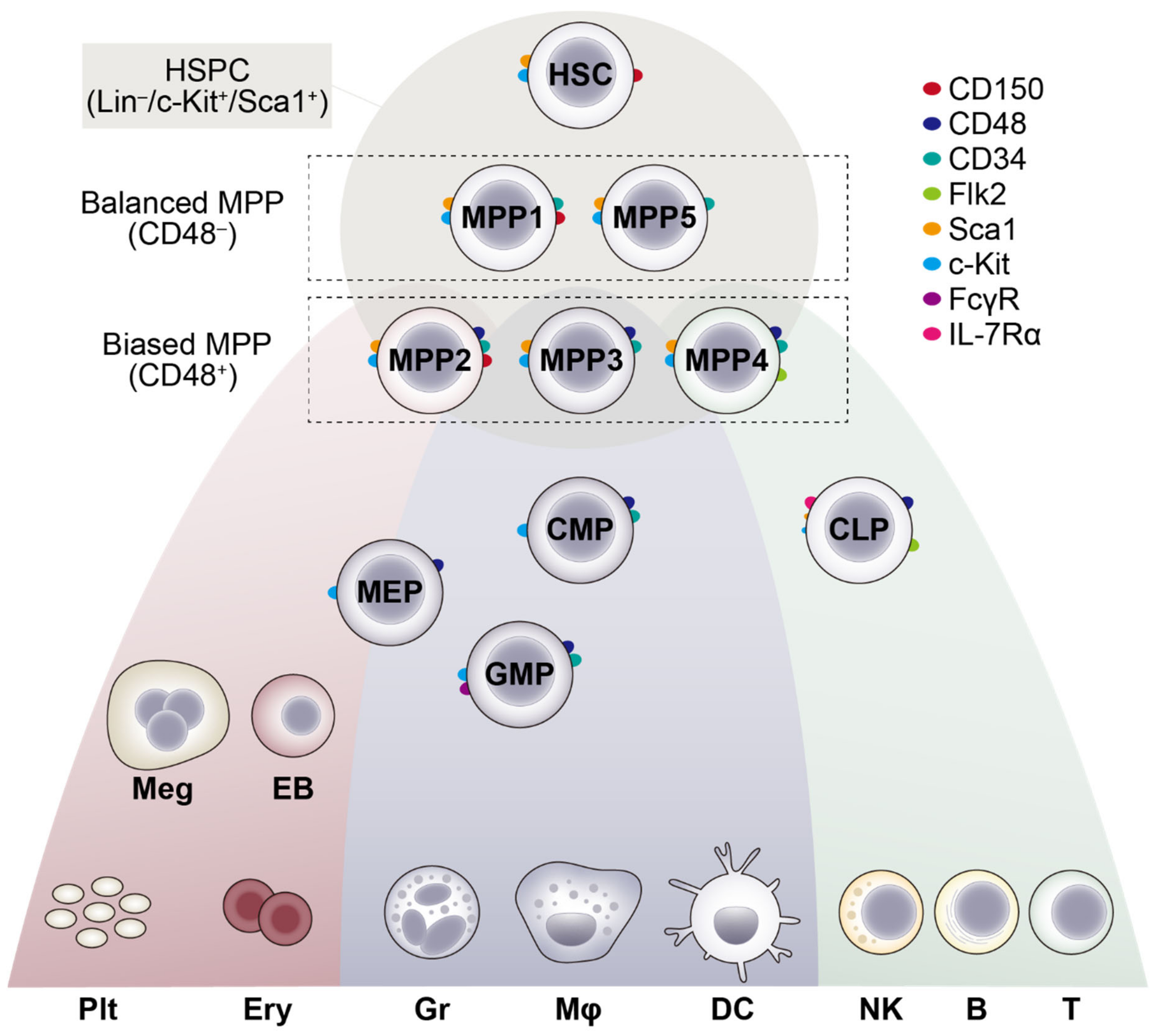
2. Hematopoietic Stem Cell Development and Clonal Expansion
3. Hematopoietic Stem Cell Clonal Behavior during Aging
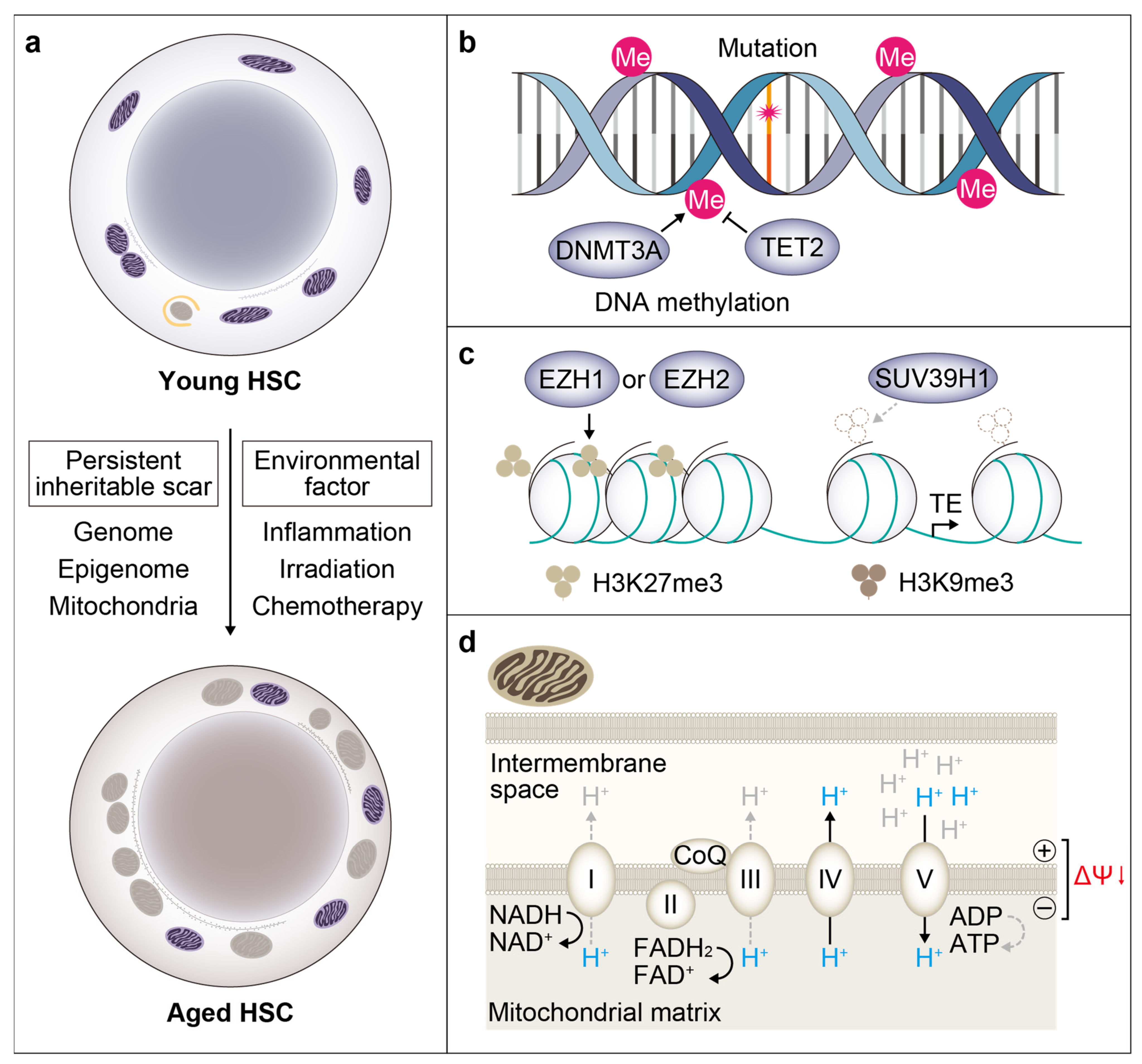
3.1. Somatic Mutation
3.2. Epigenetic Memory
References
- Orkin, S.H.; Zon, L.I. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 2008, 132, 631–644.
- Schulz, C.; Gomez Perdiguero, E.; Chorro, L.; Szabo-Rogers, H.; Cagnard, N.; Kierdorf, K.; Prinz, M.; Wu, B.; Jacobsen, S.E.; Pollard, J.W.; et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012, 336, 86–90.
- Hashimoto, D.; Chow, A.; Noizat, C.; Teo, P.; Beasley, M.B.; Leboeuf, M.; Becker, C.D.; See, P.; Price, J.; Lucas, D.; et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013, 38, 792–804.
- Beaudin, A.E.; Boyer, S.W.; Perez-Cunningham, J.; Hernandez, G.E.; Derderian, S.C.; Jujjavarapu, C.; Aaserude, E.; MacKenzie, T.; Forsberg, E.C. A Transient Developmental Hematopoietic Stem Cell Gives Rise to Innate-like B and T Cells. Cell Stem Cell 2016, 19, 768–783.
- Yamamoto, R.; Morita, Y.; Ooehara, J.; Hamanaka, S.; Onodera, M.; Rudolph, K.L.; Ema, H.; Nakauchi, H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 2013, 154, 1112–1126.
- Benz, C.; Copley, M.R.; Kent, D.G.; Wohrer, S.; Cortes, A.; Aghaeepour, N.; Ma, E.; Mader, H.; Rowe, K.; Day, C.; et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 2012, 10, 273–283.
- Pei, W.; Feyerabend, T.B.; Rossler, J.; Wang, X.; Postrach, D.; Busch, K.; Rode, I.; Klapproth, K.; Dietlein, N.; Quedenau, C.; et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 2017, 548, 456–460.
- Yamamoto, R.; Wilkinson, A.C.; Ooehara, J.; Lan, X.; Lai, C.Y.; Nakauchi, Y.; Pritchard, J.K.; Nakauchi, H. Large-Scale Clonal Analysis Resolves Aging of the Mouse Hematopoietic Stem Cell Compartment. Cell Stem Cell 2018, 22, 600–607.
- Ganuza, M.; Hall, T.; Finkelstein, D.; Wang, Y.D.; Chabot, A.; Kang, G.; Bi, W.; Wu, G.; McKinney-Freeman, S. The global clonal complexity of the murine blood system declines throughout life and after serial transplantation. Blood 2019, 133, 1927–1942.
- Ivanovs, A.; Rybtsov, S.; Ng, E.S.; Stanley, E.G.; Elefanty, A.G.; Medvinsky, A. Human haematopoietic stem cell development: From the embryo to the dish. Development 2017, 144, 2323–2337.
- Dzierzak, E.; Bigas, A. Blood Development: Hematopoietic Stem Cell Dependence and Independence. Cell Stem Cell 2018, 22, 639–651.
- Bowie, M.B.; McKnight, K.D.; Kent, D.G.; McCaffrey, L.; Hoodless, P.A.; Eaves, C.J. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J. Clin. Invest. 2006, 116, 2808–2816.
- Li, Y.; Kong, W.; Yang, W.; Patel, R.M.; Casey, E.B.; Okeyo-Owuor, T.; White, J.M.; Porter, S.N.; Morris, S.A.; Magee, J.A. Single-Cell Analysis of Neonatal HSC Ontogeny Reveals Gradual and Uncoordinated Transcriptional Reprogramming that Begins before Birth. Cell Stem Cell 2020, 27, 732–747-e737.
- Pietras, E.M.; Reynaud, D.; Kang, Y.A.; Carlin, D.; Calero-Nieto, F.J.; Leavitt, A.D.; Stuart, J.M.; Gottgens, B.; Passegue, E. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell 2015, 17, 35–46.
- Sommerkamp, P.; Romero-Mulero, M.C.; Narr, A.; Ladel, L.; Hustin, L.; Schonberger, K.; Renders, S.; Altamura, S.; Zeisberger, P.; Jacklein, K.; et al. Mouse multipotent progenitor 5 cells are located at the interphase between hematopoietic stem and progenitor cells. Blood 2021, 137, 3218–3224.
- Karamitros, D.; Stoilova, B.; Aboukhalil, Z.; Hamey, F.; Reinisch, A.; Samitsch, M.; Quek, L.; Otto, G.; Repapi, E.; Doondeea, J.; et al. Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells. Nat. Immunol. 2018, 19, 85–97.
- Velten, L.; Haas, S.F.; Raffel, S.; Blaszkiewicz, S.; Islam, S.; Hennig, B.P.; Hirche, C.; Lutz, C.; Buss, E.C.; Nowak, D.; et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 2017, 19, 271–281.
- Buenrostro, J.D.; Corces, M.R.; Lareau, C.A.; Wu, B.; Schep, A.N.; Aryee, M.J.; Majeti, R.; Chang, H.Y.; Greenleaf, W.J. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 2018, 173, 1535–1548.
- Dahlin, J.S.; Hamey, F.K.; Pijuan-Sala, B.; Shepherd, M.; Lau, W.W.Y.; Nestorowa, S.; Weinreb, C.; Wolock, S.; Hannah, R.; Diamanti, E.; et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood 2018, 131, e1–e11.
- Pellin, D.; Loperfido, M.; Baricordi, C.; Wolock, S.L.; Montepeloso, A.; Weinberg, O.K.; Biffi, A.; Klein, A.M.; Biasco, L. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat. Commun. 2019, 10, 2395.
- Challen, G.A.; Pietras, E.M.; Wallscheid, N.C.; Signer, R.A.J. Simplified murine multipotent progenitor isolation scheme: Establishing a consensus approach for multipotent progenitor identification. Exp. Hematol. 2021, 104, 55–63.
- Kobayashi, H.; Morikawa, T.; Okinaga, A.; Hamano, F.; Hashidate-Yoshida, T.; Watanuki, S.; Hishikawa, D.; Shindou, H.; Arai, F.; Kabe, Y.; et al. Environmental Optimization Enables Maintenance of Quiescent Hematopoietic Stem Cells Ex Vivo. Cell Rep. 2019, 28, 145–158.
- Wilson, A.; Laurenti, E.; Oser, G.; van der Wath, R.C.; Blanco-Bose, W.; Jaworski, M.; Offner, S.; Dunant, C.F.; Eshkind, L.; Bockamp, E.; et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 2008, 135, 1118–1129.
- Cheshier, S.H.; Prohaska, S.S.; Weissman, I.L. The effect of bleeding on hematopoietic stem cell cycling and self-renewal. Stem Cells Dev. 2007, 16, 707–717.
- Beerman, I.; Bock, C.; Garrison, B.S.; Smith, Z.D.; Gu, H.; Meissner, A.; Rossi, D.J. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell 2013, 12, 413–425.
- Foudi, A.; Hochedlinger, K.; Van Buren, D.; Schindler, J.W.; Jaenisch, R.; Carey, V.; Hock, H. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 2009, 27, 84–90.
- Bernitz, J.M.; Kim, H.S.; MacArthur, B.; Sieburg, H.; Moore, K. Hematopoietic Stem Cells Count and Remember Self-Renewal Divisions. Cell 2016, 167, 1296–1309 e1210.
- Bernitz, J.M.; Rapp, K.; Daniel, M.G.; Shcherbinin, D.; Yuan, Y.; Gomes, A.; Waghray, A.; Brosh, R.; Lachmann, A.; Ma’ayan, A.; et al. Memory of Divisional History Directs the Continuous Process of Primitive Hematopoietic Lineage Commitment. Stem Cell Rep. 2020, 14, 561–574.
- Challen, G.A.; Goodell, M.A. Promiscuous expression of H2B-GFP transgene in hematopoietic stem cells. PLoS ONE 2008, 3, e2357.
- Morcos, M.N.F.; Zerjatke, T.; Glauche, I.; Munz, C.M.; Ge, Y.; Petzold, A.; Reinhardt, S.; Dahl, A.; Anstee, N.S.; Bogeska, R.; et al. Continuous mitotic activity of primitive hematopoietic stem cells in adult mice. J. Exp. Med. 2020, 217, e20191284.
- Grover, A.; Sanjuan-Pla, A.; Thongjuea, S.; Carrelha, J.; Giustacchini, A.; Gambardella, A.; Macaulay, I.; Mancini, E.; Luis, T.C.; Mead, A.; et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 2016, 7, 11075.
- Morrison, S.J.; Wandycz, A.M.; Akashi, K.; Globerson, A.; Weissman, I.L. The aging of hematopoietic stem cells. Nat. Med. 1996, 2, 1011–1016.
- Dykstra, B.; Olthof, S.; Schreuder, J.; Ritsema, M.; de Haan, G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 2011, 208, 2691–2703.
- Rossi, D.J.; Bryder, D.; Zahn, J.M.; Ahlenius, H.; Sonu, R.; Wagers, A.J.; Weissman, I.L. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA 2005, 102, 9194–9199.
- Busque, L.; Mio, R.; Mattioli, J.; Brais, E.; Blais, N.; Lalonde, Y.; Maragh, M.; Gilliland, D.G. Nonrandom X-inactivation patterns in normal females: Lyonization ratios vary with age. Blood 1996, 88, 59–65.
- Sawen, P.; Eldeeb, M.; Erlandsson, E.; Kristiansen, T.A.; Laterza, C.; Kokaia, Z.; Karlsson, G.; Yuan, J.; Soneji, S.; Mandal, P.K.; et al. Murine HSCs contribute actively to native hematopoiesis but with reduced differentiation capacity upon aging. eLife 2018, 7.
- Herault, L.; Poplineau, M.; Mazuel, A.; Platet, N.; Remy, E.; Duprez, E. Single-cell RNA-seq reveals a concomitant delay in differentiation and cell cycle of aged hematopoietic stem cells. BMC Biol. 2021, 19, 19.
- Mann, M.; Mehta, A.; de Boer, C.G.; Kowalczyk, M.S.; Lee, K.; Haldeman, P.; Rogel, N.; Knecht, A.R.; Farouq, D.; Regev, A.; et al. Heterogeneous Responses of Hematopoietic Stem Cells to Inflammatory Stimuli Are Altered with Age. Cell Rep. 2018, 25, 2992–3005 e2995.
- Kowalczyk, M.S.; Tirosh, I.; Heckl, D.; Rao, T.N.; Dixit, A.; Haas, B.J.; Schneider, R.K.; Wagers, A.J.; Ebert, B.L.; Regev, A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015, 25, 1860–1872.
- Ho, T.T.; Warr, M.R.; Adelman, E.R.; Lansinger, O.M.; Flach, J.; Verovskaya, E.V.; Figueroa, M.E.; Passegue, E. Autophagy maintains the metabolism and function of young and old stem cells. Nature 2017, 543, 205–210.
- Mansell, E.; Sigurdsson, V.; Deltcheva, E.; Brown, J.; James, C.; Miharada, K.; Soneji, S.; Larsson, J.; Enver, T. Mitochondrial Potentiation Ameliorates Age-Related Heterogeneity in Hematopoietic Stem Cell Function. Cell Stem Cell 2020.
- Lengefeld, J.; Cheng, C.W.; Maretich, P.; Blair, M.; Hagen, H.; McReynolds, M.R.; Sullivan, E.; Majors, K.; Roberts, C.; Kang, J.H.; et al. Cell size is a determinant of stem cell potential during aging. Sci. Adv. 2021, 7, eabk0271.
- Osorio, F.G.; Rosendahl Huber, A.; Oka, R.; Verheul, M.; Patel, S.H.; Hasaart, K.; de la Fonteijne, L.; Varela, I.; Camargo, F.D.; van Boxtel, R. Somatic Mutations Reveal Lineage Relationships and Age-Related Mutagenesis in Human Hematopoiesis. Cell Rep. 2018, 25, 2308–2316.
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; Miller, C.A.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478.
- Genovese, G.; Kahler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487.
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498.
- Challen, G.A.; Sun, D.; Jeong, M.; Luo, M.; Jelinek, J.; Berg, J.S.; Bock, C.; Vasanthakumar, A.; Gu, H.; Xi, Y.; et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011, 44, 23–31.
- Koya, J.; Kataoka, K.; Sato, T.; Bando, M.; Kato, Y.; Tsuruta-Kishino, T.; Kobayashi, H.; Narukawa, K.; Miyoshi, H.; Shirahige, K.; et al. DNMT3A R882 mutants interact with polycomb proteins to block haematopoietic stem and leukaemic cell differentiation. Nat. Commun. 2016, 7, 10924.
- Moran-Crusio, K.; Reavie, L.; Shih, A.; Abdel-Wahab, O.; Ndiaye-Lobry, D.; Lobry, C.; Figueroa, M.E.; Vasanthakumar, A.; Patel, J.; Zhao, X.; et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011, 20, 11–24.
- Quivoron, C.; Couronne, L.; Della Valle, V.; Lopez, C.K.; Plo, I.; Wagner-Ballon, O.; Do Cruzeiro, M.; Delhommeau, F.; Arnulf, B.; Stern, M.H.; et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011, 20, 25–38.
- Fujino, T.; Goyama, S.; Sugiura, Y.; Inoue, D.; Asada, S.; Yamasaki, S.; Matsumoto, A.; Yamaguchi, K.; Isobe, Y.; Tsuchiya, A.; et al. Mutant ASXL1 induces age-related expansion of phenotypic hematopoietic stem cells through activation of Akt/mTOR pathway. Nat. Commun. 2021, 12, 1826.
- Lundberg, P.; Takizawa, H.; Kubovcakova, L.; Guo, G.; Hao-Shen, H.; Dirnhofer, S.; Orkin, S.H.; Manz, M.G.; Skoda, R.C. Myeloproliferative neoplasms can be initiated from a single hematopoietic stem cell expressing JAK2-V617F. J. Exp. Med. 2014, 211, 2213–2230.
- Mullally, A.; Lane, S.W.; Ball, B.; Megerdichian, C.; Okabe, R.; Al-Shahrour, F.; Paktinat, M.; Haydu, J.E.; Housman, E.; Lord, A.M.; et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 2010, 17, 584–596.
- Liu, Y.; Elf, S.E.; Miyata, Y.; Sashida, G.; Liu, Y.; Huang, G.; Di Giandomenico, S.; Lee, J.M.; Deblasio, A.; Menendez, S.; et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 2009, 4, 37–48.
- Bondar, T.; Medzhitov, R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell 2010, 6, 309–322.
- Hsu, J.I.; Dayaram, T.; Tovy, A.; De Braekeleer, E.; Jeong, M.; Wang, F.; Zhang, J.; Heffernan, T.P.; Gera, S.; Kovacs, J.J.; et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell 2018, 23, 700–713.
- Kahn, J.D.; Miller, P.G.; Silver, A.J.; Sellar, R.S.; Bhatt, S.; Gibson, C.; McConkey, M.; Adams, D.; Mar, B.; Mertins, P.; et al. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood 2018, 132, 1095–1105.
- Jaiswal, S.; Natarajan, P.; Silver, A.J.; Gibson, C.J.; Bick, A.G.; Shvartz, E.; McConkey, M.; Gupta, N.; Gabriel, S.; Ardissino, D.; et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl J. Med. 2017, 377, 111–121.
- Kohnke, T.; Majeti, R. Clonal Hematopoiesis: From Mechanisms to Clinical Intervention. Cancer Discov. 2021.
- Rundberg Nilsson, A.; Soneji, S.; Adolfsson, S.; Bryder, D.; Pronk, C.J. Human and Murine Hematopoietic Stem Cell Aging Is Associated with Functional Impairments and Intrinsic Megakaryocytic/Erythroid Bias. PLoS ONE 2016, 11, e0158369.
- Stewart-Morgan, K.R.; Petryk, N.; Groth, A. Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 2020, 22, 361–371.
- Iwama, A.; Oguro, H.; Negishi, M.; Kato, Y.; Morita, Y.; Tsukui, H.; Ema, H.; Kamijo, T.; Katoh-Fukui, Y.; Koseki, H.; et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 2004, 21, 843–851.
- Trowbridge, J.J.; Snow, J.W.; Kim, J.; Orkin, S.H. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 2009, 5, 442–449.
- Kuribayashi, W.; Oshima, M.; Itokawa, N.; Koide, S.; Nakajima-Takagi, Y.; Yamashita, M.; Yamazaki, S.; Rahmutulla, B.; Miura, F.; Ito, T.; et al. Limited rejuvenation of aged hematopoietic stem cells in young bone marrow niche. J. Exp. Med. 2021, 218.
- Ho, T.T.; Dellorusso, P.V.; Verovskaya, E.V.; Bakker, S.T.; Flach, J.; Smith, L.K.; Ventura, P.B.; Lansinger, O.M.; Herault, A.; Zhang, S.Y.; et al. Aged hematopoietic stem cells are refractory to bloodborne systemic rejuvenation interventions. J. Exp. Med. 2021, 218, e20210223.
- Sun, D.; Luo, M.; Jeong, M.; Rodriguez, B.; Xia, Z.; Hannah, R.; Wang, H.; Le, T.; Faull, K.F.; Chen, R.; et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 2014, 14, 673–688.
- Adelman, E.R.; Huang, H.T.; Roisman, A.; Olsson, A.; Colaprico, A.; Qin, T.; Lindsley, R.C.; Bejar, R.; Salomonis, N.; Grimes, H.L.; et al. Aging Human Hematopoietic Stem Cells Manifest Profound Epigenetic Reprogramming of Enhancers That May Predispose to Leukemia. Cancer Discov. 2019, 9, 1080–1101.
- Hidalgo, I.; Herrera-Merchan, A.; Ligos, J.M.; Carramolino, L.; Nunez, J.; Martinez, F.; Dominguez, O.; Torres, M.; Gonzalez, S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell 2012, 11, 649–662.
- Flohr Svendsen, A.; Yang, D.; Kim, K.; Lazare, S.; Skinder, N.; Zwart, E.; Mura-Meszaros, A.; Ausema, A.; von Eyss, B.; de Haan, G.; et al. A comprehensive transcriptome signature of murine hematopoietic stem cell aging. Blood 2021, 138, 439–451.
- Mochizuki-Kashio, M.; Aoyama, K.; Sashida, G.; Oshima, M.; Tomioka, T.; Muto, T.; Wang, C.; Iwama, A. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood 2015, 126, 1172–1183.
- Xie, H.; Xu, J.; Hsu, J.H.; Nguyen, M.; Fujiwara, Y.; Peng, C.; Orkin, S.H. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 2014, 14, 68–80.
- Djeghloul, D.; Kuranda, K.; Kuzniak, I.; Barbieri, D.; Naguibneva, I.; Choisy, C.; Bories, J.C.; Dosquet, C.; Pla, M.; Vanneaux, V.; et al. Age-Associated Decrease of the Histone Methyltransferase SUV39H1 in HSC Perturbs Heterochromatin and B Lymphoid Differentiation. Stem Cell Rep. 2016, 6, 970–984.
- Clapes, T.; Polyzou, A.; Prater, P.; Sagar; Morales-Hernandez, A.; Ferrarini, M.G.; Kehrer, N.; Lefkopoulos, S.; Bergo, V.; Hummel, B.; et al. Chemotherapy-induced transposable elements activate MDA5 to enhance haematopoietic regeneration. Nat. Cell Biol. 2021, 23, 704–717.
- Ghafouri-Fard, S.; Niazi, V.; Taheri, M. Role of miRNAs and lncRNAs in hematopoietic stem cell differentiation. Noncoding RNA Res. 2021, 6, 8–14.
- Mehta, A.; Zhao, J.L.; Sinha, N.; Marinov, G.K.; Mann, M.; Kowalczyk, M.S.; Galimidi, R.P.; Du, X.; Erikci, E.; Regev, A.; et al. The MicroRNA-132 and MicroRNA-212 Cluster Regulates Hematopoietic Stem Cell Maintenance and Survival with Age by Buffering FOXO3 Expression. Immunity 2015, 42, 1021–1032.
- Grants, J.M.; Wegrzyn, J.; Hui, T.; O’Neill, K.; Shadbolt, M.; Knapp, D.; Parker, J.; Deng, Y.; Gopal, A.; Docking, T.R.; et al. Altered microRNA expression links IL6 and TNF-induced inflammaging with myeloid malignancy in humans and mice. Blood 2020, 135, 2235–2251.
- Luo, M.; Jeong, M.; Sun, D.; Park, H.J.; Rodriguez, B.A.; Xia, Z.; Yang, L.; Zhang, X.; Sheng, K.; Darlington, G.J.; et al. Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell 2015, 16, 426–438.
- Ranzoni, A.M.; Tangherloni, A.; Berest, I.; Riva, S.G.; Myers, B.; Strzelecka, P.M.; Xu, J.; Panada, E.; Mohorianu, I.; Zaugg, J.B.; et al. Integrative Single-Cell RNA-Seq and ATAC-Seq Analysis of Human Developmental Hematopoiesis. Cell Stem Cell 2021, 28, 472–487-e477.




