
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Harley Myler | + 2268 word(s) | 2268 | 2021-12-22 04:37:16 | | | |
| 2 | Peter Tang | Meta information modification | 2268 | 2022-01-08 08:04:24 | | | | |
| 3 | Lindsay Dong | Meta information modification | 2268 | 2022-03-28 08:11:57 | | |
Video Upload Options
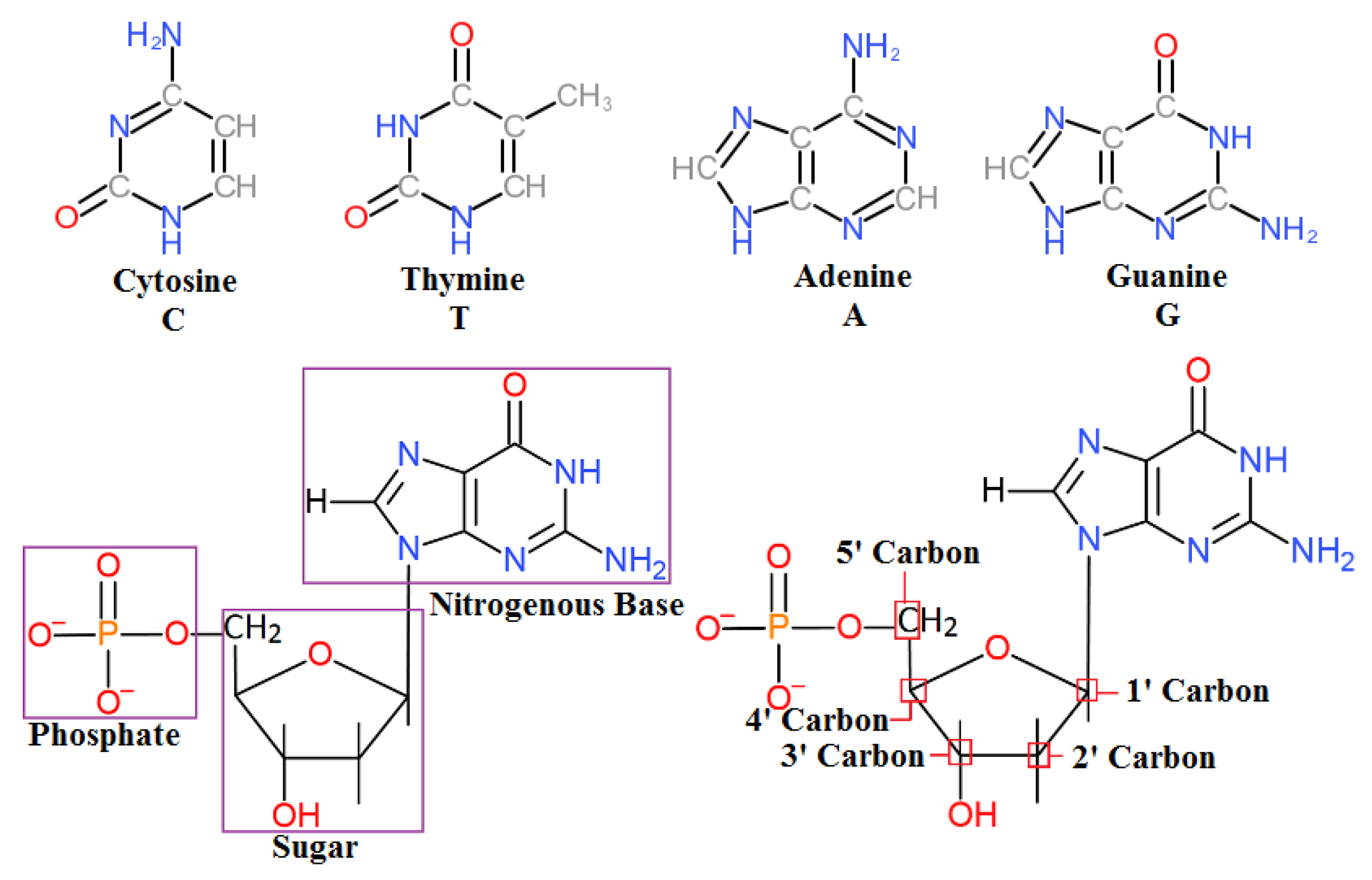
DNA sequencing is a critical functionality in biomedical research, and technical advances that improve it have important implications for human health. The overall structure of DNA is negatively charged and has a negative electrostatic potential due to a negatively charged phosphate backbone. Nucleotides are electroactive compounds that produce reduction and oxidation signals after hybridization. These high electrostatic potentials can be exploited with VLSI charge-sensitive electronic structures. The sequence of DNA is encoded within each nitrogenous base: adenine (A), guanine (G), cytosine (C), or thymine (T), and this 4-bit system allows for large amounts of information to be stored in a single DNA molecule, which may contain upward of 3 billion bases.
1. Introduction
2. Limitations of Existing Technologies and Need for VLSI Biosensors for Sequencing
|
Sequencing Method |
Industry |
Limitation |
|---|---|---|
|
Ion semiconductor |
Life Technologies |
Homopolymer errors [10] |
|
454 Pyrosequencing |
ROCHE |
Expensive and homopolymer errors [11] |
|
Sequencing by synthesis (cyclic reversible termination) |
Illumina, Qiagen |
Expensive equipment [12] |
|
Sequencing by ligation |
Applied Biosystems (SOLiD) |
Very slow, unable to read palindromic regions [13] |
|
Sequencing Method |
Industry |
Limitation |
|---|---|---|
|
Single-molecule real-time |
Pacific Bioscience |
Higher error rate, Low throughput, Expensive equipment [14] |
|
Nanopore |
ONT |
Large error rate, Limitation on homopolymer sequencing [15] |
3. VLSI Architectures in DNA Detection
3.1. Electrolyte-Insulator Silicon (EIS) Capacitor
3.2. TFT (Thin Film Transistor)
3.3. Different Types of FET’s
3.3.1. MOSFET (Metal Oxide Semiconductor FET)
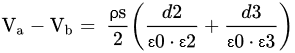
3.3.2. CMFET (Charge Modulated FET)
3.3.3. ISFET (Ion Sensitive FET)
3.4. NWFET (Nanowire FET)
3.5. Carbon Nanotube FET (CNTFET)
3.6. Graphene FET
3.7. CMOS (Complementary Metal Oxide Semiconductor)
3.8. Nanopore FET
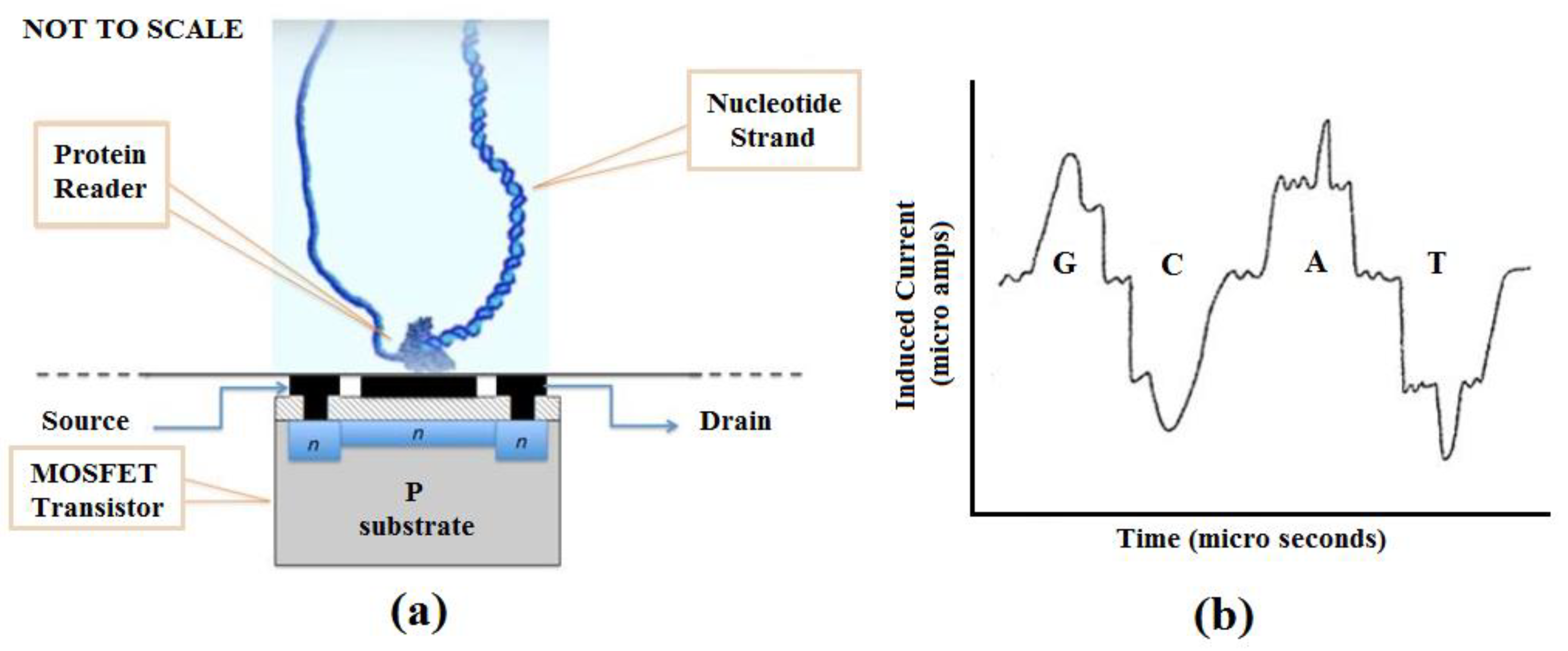
4. A Novel Method for DNA Sequencing
References
- Rohs, R.; Jin, X.; West, S.; Joshi, R.; Honig, B.; Mann, R.S. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010, 79, 233–269.
- Lipfert, J.; Doniach, S.; Das, R.; Herschlag, D. Understanding nucleic acid-ion interactions. Annu. Rev. Biochem. 2014, 83, 813–841.
- Weiner, P.K.; Langridge, R.; Blaney, J.M.; Schaefer, R.; Kollman, P.A. Electrostatic potential molecular surfaces. Proc. Natl. Acad. Sci. USA 1982, 79, 3754–3758.
- Watson, J.D.; Crick, F.H.C. Reprint: Molecular structure of nucleic acids. Ann. Intern. Med. 2003, 138, 581.
- Frank-Kamenetskii, M.D.; Anshelevich, V.V.; Lukashin, A.V. Polyelectrolyte model of DNA. Uspekhi Fiz. Nauk 1987, 151, 595.
- Grosberg, A.Y.; Nguyen, T.T.; Shklovskii, B.I. Colloquium: The physics of charge inversion in chemical and biological systems. Rev. Mod. Phys. 2002, 74, 329–345.
- Paleček, E. Oscillographic polarography of highly polymerized Deoxyribonucleic acid. Nature 1960, 188, 656–657.
- Skogerboe, K.J. Molecular biology techniques. Anal. Chem. 1995, 67, 449–454.
- Men, A.E.; Wilson, P.; Siemering, K.; Forrest, S. Sanger DNA sequencing. In Next Generation Genome Sequencing: Towards Personalized Medicine; Wiley-VCH: Weinheim, Germany, 2008; ISBN 9783527320905.
- Feng, W.; Zhao, S.; Xue, D.; Song, F.; Li, Z.; Chen, D.; He, B.; Hao, Y.; Wang, Y.; Liu, Y. Improving alignment accuracy on homopolymer regions for semiconductor-based sequencing technologies. BMC Genom. 2016, 17, 521.
- Ronaghi, M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001, 11, 3–11.
- Kircher, M.; Kelso, J. High-throughput DNA sequencing - Concepts and limitations. BioEssays 2010, 32, 524–536.
- Huang, Y.F.; Chen, S.C.; Chiang, Y.S.; Chen, T.H.; Chiu, K.P. Palindromic sequence impedes sequencing-by-ligation mechanism. BMC Syst. Biol. 2012, 6, S10.
- Rhoads, A.; Au, K.F. PacBio Sequencing and its applications. Genom. Proteom. Bioinforma 2015, 13, 278–289.
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351.
- Magierowski, S.; Huang, Y.; Wang, C.; Ghafar-Zadeh, E. Nanopore-CMOS interfaces for DNA sequencing. Biosensors 2016, 6, 42.
- Cooper, E.B.; Fritz, J.; Wiegand, G.; Wagner, P.; Manalis, S.R. Robust microfabricated field-effect sensor for monitoring molecular adsorption in liquids. Appl. Phys. Lett. 2001, 79, 3875–3877.
- Fritz, J.; Cooper, E.B.; Gaudet, S.; Sorger, P.K.; Manalis, S.R. Electronic detection of DNA by its intrinsic molecular charge. Proc. Natl. Acad. Sci. USA 2002, 99, 14142–14146.
- Estrela, P.; Stewart, A.G.; Migliorato, P.; Maeda, H. Label-free detection of DNA hybridization with Au/SiO2/Si diodes and poly-Si TFTs. In Proceedings of the Technical Digest—International Electron Devices Meeting, San Francisco, CA, USA, 13–15 December 2004.
- Kim, D.S.; Jeong, Y.T.; Park, H.J.; Shin, J.K.; Choi, P.; Lee, J.H.; Lim, G. An FET-type charge sensor for highly sensitive detection of DNA sequence. Biosens. Bioelectron. 2004, 20, 69–74.
- Barbaro, M.; Bonfiglio, A.; Raffo, L. A charge-modulated FET for detection of biomolecular processes: Conception, modeling, and simulation. IEEE Trans. Electron Devices 2006, 53, 158–166.
- De Lumley-Woodyear, T.; Campbell, C.N.; Freeman, E.; Freeman, A.; Georgiou, G.; Heller, A. Rapid amperometric verification of PCR amplification of DNA. Anal. Chem. 1999, 71, 535–538.
- Bergveld, P. Short communications: development of an ion-sensitive solid-state device for neurophysiological measurements. IEEE Trans. Biomed. Eng. 1970, 70–71.
- Zhang, A.; Zheng, G.; Lieber, C. Nanowires : Building Blocks for Nanoscience and Nanotechnology; Springer International Publishing: Cham, Switzerland, 2016; ISBN 9783319419817.
- Cui, Y.; Duan, X.; Hu, J.; Lieber, C.M. Doping and electrical transport in silicon nanowires. J. Phys. Chem. B 2000, 104, 5213–5216.
- Snow, E.S.; Perkins, F.K.; Houser, E.J.; Badescu, S.C.; Reinecke, T.L. Chemical detection with a single-walled carbon nanotube capacitor. Science 2005, 307, 1942–1945.
- Mehrabani, S.; Maker, A.J.; Armani, A.M. Hybrid integrated label-free chemical and biological sensors. Sensors 2014, 14, 5890–5928.
- Daniel, S.; Rao, T.P.; Rao, K.S.; Rani, S.U.; Naidu, G.R.K.; Lee, H.Y.; Kawai, T. A review of DNA functionalized/grafted carbon nanotubes and their characterization. Sens. Actuators B Chem. 2007, 122, 672–682.
- Hu, P.A.; Zhang, J.; Li, L.; Wang, Z.; O’Neill, W.; Estrela, P. Carbon nanostructure-based field-effect transistors for label-free chemical/biological sensors. Sensors 2010, 10, 5133–5159.
- Michael, T.H.; Preston, B.L.; Joon, L.; Duyoung, C.; Alexander, H.M.; Gennadi, G.; Ratnesh, L. Highly specific SNP detection using 2D graphene electronics and DNA strand displacement. Proc. Natl. Acad. Sci. USA 2016, 13, 7088–7093.
- Lai, S.; Caboni, A.; Loi, D.; Barbaro, M. A CMOS biocompatible charge detector for biosensing applications. IEEE Trans. Electron Devices 2012, 59, 2512–2519.
- Xie, P.; Xiong, Q.; Fang, Y.; Qing, Q.; Lieber, C.M. Local electrical potential detection of DNA by nanowire-nanopore sensors. Nat. Nanotechnol. 2011, 7, 119–125.
- Bedell, S.; D’Emic, C.; Peng, H.; Zafar, S. FET Nanopore Sensor. U.S. Patent US8828138B2, 9 September 2014.
- Moore, I.; Millar, C.; Roy, S.; Asenov, A. FET based nano-pore sensing: A 3D simulation study. J. Comput. Electron. 2012, 11, 266–271.
- Sauer, J.; Zeghbroeck, J. Method and Apparatus for Detecting Nucleotides. U.S. Patent 20130264204A9, 10 October 2013.




