
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mauro Mandrioli | + 2608 word(s) | 2608 | 2021-12-28 03:00:37 | | | |
| 2 | Vivi Li | Meta information modification | 2608 | 2021-12-31 02:23:19 | | |
Video Upload Options
In the last decade, genome editing technologies became very effective and several clinical trials have been started in order to use them for treating some genetic diseases. Interestingly, despite more than 50 years of discussion about the frontiers of genetics in human health and evolution, the debate about the bioethics and the regulatory practices of genome editing is still far from satisfactory answers. This delay results from an excessive emphasis on the effectiveness of the genome editing technologies that is relevant for the regulatory practices, but not at a bioethical level. Indeed, other factors (such as accessibility and acceptability) could make these techniques not accepted at the bioethical level, even in the presence of their 100% effectiveness.
1. Introduction
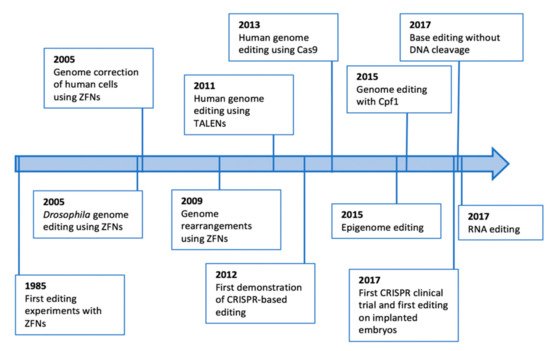
2. Origin, Promises and Pitfalls of the Genome Editing Technologies
3. The Role of Policy Makers and Bioethicists in the Establishment of the Limits in Genome Editing
4. How Do We Begin to Regulate Genome Editing Technologies?
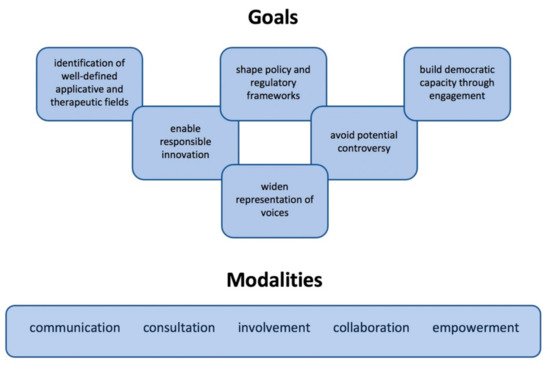
References
- Karen Bulaklak; Charles A. Gersbach; The once and future gene therapy. Nature Communications 2020, 11, 1-4, 10.1038/s41467-020-19505-2.
- Asher Mullard; Gene-editing pipeline takes off. Nature Reviews Drug Discovery 2020, 19, 367-372, 10.1038/d41573-020-00096-y.
- Jonathan D. Finn; Amy Rhoden Smith; Mihir C. Patel; Lucinda Shaw; Madeleine R. Youniss; Jane Van Heteren; Tanner Dirstine; Corey Ciullo; Reynald Lescarbeau; Jessica Seitzer; et al.Ruchi R. ShahAalok ShahDandan LingJacqueline GroweMelissa PinkEllen RohdeKristy M. WoodWilliam E. SalomonWilliam F. HarringtonChristian DombrowskiWalter R. StrappsYong ChangDavid V. Morrissey A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Reports 2018, 22, 2227-2235, 10.1016/j.celrep.2018.02.014.
- Edward A. Stadtmauer; Joseph A. Fraietta; Megan M. Davis; Adam D. Cohen; Kristy L. Weber; Eric Lancaster; Patricia A. Mangan; Irina Kulikovskaya; Minnal Gupta; Fang Chen; et al.Lifeng TianVanessa E. GonzalezJun XuIn-Young JungJ. Joseph MelenhorstGabriela PlesaJoanne SheaTina MatlawskiAmanda CerviniAvery L. GaymonStephanie DesjardinsAnne LamontagneJanuary Salas-MckeeAndrew FesnakDonald L. SiegelBruce L. LevineJulie K. JadlowskyRegina M. YoungAnne ChewWei-Ting HwangElizabeth O. HexnerBeatriz M. CarrenoChristopher L. NoblesFrederic D. BushmanKevin R. ParkerYanyan QiAnsuman T. SatpathyHoward Y. ChangYangbing ZhaoSimon F. LaceyCarl H. June CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367, eaba736, 10.1126/science.aba7365.
- Adrian Pickar-Oliver; Charles A. Gersbach; The next generation of CRISPR–Cas technologies and applications. Nature Reviews Molecular Cell Biology 2019, 20, 490-507, 10.1038/s41580-019-0131-5.
- Andrew V. Anzalone; Luke Koblan; David R. Liu; Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nature Biotechnology 2020, 38, 824-844, 10.1038/s41587-020-0561-9.
- Mark Thomas; Gaetan Burgio; David J. Adams; Vivek Iyer; Collateral damage and CRISPR genome editing. PLOS Genetics 2019, 15, e1007994, 10.1371/journal.pgen.1007994.
- Ryan J. Marina; Kristopher W. Brannan; Kevin D. Dong; Brian A. Yee; Gene W. Yeo; Evaluation of Engineered CRISPR-Cas-Mediated Systems for Site-Specific RNA Editing. Cell Reports 2020, 33, 108350, 10.1016/j.celrep.2020.108350.
- Ahsen Özcan; Rohan Krajeski; Eleonora Ioannidi; Brennan Lee; Apolonia Gardner; Kira S. Makarova; Eugene V. Koonin; Omar O. Abudayyeh; Jonathan S. Gootenberg; Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature 2021, 597, 720-725, 10.1038/s41586-021-03886-5.
- Pratiksha I. Thakore; Joshua B. Black; Isaac B. Hilton; Charles A. Gersbach; Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nature Methods 2016, 13, 127-137, 10.1038/nmeth.3733.
- Rutger A.F. Gjaltema; Marianne G. Rots; Advances of epigenetic editing. Current Opinion in Chemical Biology 2020, 57, 75-81, 10.1016/j.cbpa.2020.04.020.
- Muneaki Nakamura; Yuchen Gao; Antonia A. Dominguez; Lei S. Qi; CRISPR technologies for precise epigenome editing. Nature Cell Biology 2021, 23, 11-22, 10.1038/s41556-020-00620-7.
- Heidi Ledford; Beyond CRISPR: A guide to the many other ways to edit a genome. Nature 2016, 536, 137-137, 10.1038/536136b.
- Bogdan Kirillov; Ekaterina Savitskaya; Maxim Panov; Aleksey Y Ogurtsov; Svetlana A Shabalina; Eugene V Koonin; Konstantin V Severinov; Uncertainty-aware and interpretable evaluation of Cas9–gRNA and Cas12a–gRNA specificity for fully matched and partially mismatched targets with Deep Kernel Learning. Nucleic Acids Research 2021, 12, gkab1065, 10.1093/nar/gkab1065.
- Omar O. Abudayyeh; Jonathan S. Gootenberg; Patrick Essletzbichler; Shuo Han; Julia Joung; Joseph J. Belanto; Vanessa Verdine; David B. T. Cox; Max J. Kellner; Aviv Regev; et al.Eric S. LanderDaniel VoytasAlice Y. TingFeng Zhang RNA targeting with CRISPR–Cas13. Nature 2017, 550, 280-284, 10.1038/nature24049.
- Rodrigo A. Gier; Krista A. Budinich; Niklaus H. Evitt; Zhendong Cao; Elizabeth S. Freilich; Qingzhou Chen; Jun Qi; Yemin Lan; Rahul M. Kohli; Junwei Shi; et al. High-performance CRISPR-Cas12a genome editing for combinatorial genetic screening. Nature Communications 2020, 11, 1-9, 10.1038/s41467-020-17209-1.
- Kira S. Makarova; Yuri I. Wolf; Jaime Iranzo; Sergey A. Shmakov; Omer S. Alkhnbashi; Stan Brouns; Emmanuelle Charpentier; David Cheng; Daniel H. Haft; Philippe Horvath; et al.Sylvain MoineauFrancisco J. M. MojicaDavid ScottShiraz A. ShahVirginijus SiksnysMichael P. TernsČeslovas VenclovasMalcolm F. WhiteAlexander F. YakuninWinston YanFeng ZhangRoger A. GarrettRolf BackofenJohn Van Der OostRodolphe BarrangouEugene V. Koonin Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nature Reviews Genetics 2019, 18, 67-83, 10.1038/s41579-019-0299-x.
- Carina Edmondson; Qi Zhou; Xuan Liu; Analysis of conventional and alternative CRISPR/Cas9 genome editing to enhance a single-base pair knock-in mutation. BMC Biotechnology 2021, 21, 1-9, 10.1186/s12896-021-00707-5.
- S. Scherer; R. W. Davis; Replacement of chromosome segments with altered DNA sequences constructed in vitro.. Proceedings of the National Academy of Sciences 1979, 76, 4951-4955, 10.1073/pnas.76.10.4951.
- Kirk R. Thomas; Kim R. Folger; Mario R. Capecchi; High frequency targeting of genes to specific sites in the mammalian genome. Cell 1986, 44, 419-428, 10.1016/0092-8674(86)90463-0.
- Ross Chapman; Martin R.G. Taylor; Simon J. Boulton; Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Molecular Cell 2012, 47, 497-510, 10.1016/j.molcel.2012.07.029.
- Y Ishino; H Shinagawa; K Makino; M Amemura; A Nakata; Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology 1987, 169, 5429-5433, 10.1128/jb.169.12.5429-5433.1987.
- Ruud. Jansen; Jan. D. A. Van Embden; Wim. Gaastra; Leo. M. Schouls; Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology 2002, 43, 1565-1575, 10.1046/j.1365-2958.2002.02839.x.
- Francisco J. M. Mojica; Elena Soria; Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. Journal of Molecular Evolution 2005, 60, 174-182, 10.1007/s00239-004-0046-3.
- Alexander Bolotin; Benoit Quinquis; Alexei Sorokine; S. Dusko Ehrlich; Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551-2561, 10.1099/mic.0.28048-0.
- Rodolphe Barrangou; Christophe Fremaux; Hélène Deveau; Melissa Richards; Patrick Boyaval; Sylvain Moineau; Dennis A. Romero; Philippe Horvath; CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709-1712, 10.1126/science.1138140.
- Martin Jinek; Krzysztof Chylinski; Ines Fonfara; Michael Hauer; Jennifer A. Doudna; Emmanuelle Charpentier; A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816-821, 10.1126/science.1225829.
- Benjamin Davies; The technical risks of human gene editing. Human Reproduction 2019, 34, 2104-2111, 10.1093/humrep/dez162.
- Thomas Gaj; Charles A. Gersbach; Carlos F. Barbas; ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology 2013, 31, 397-405, 10.1016/j.tibtech.2013.04.004.
- Xiao-Hui Zhang; Louis Y Tee; Xiao-Gang Wang; Qun-Shan Huang; Shi-Hua Yang; Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy - Nucleic Acids 2015, 4, e264, 10.1038/mtna.2015.37.
- Gregorio Alanis-Lobato; Jasmin Zohren; Afshan McCarthy; Norah M. E. Fogarty; Nada Kubikova; Emily Hardman; Maria Greco; Dagan Wells; James M. A. Turner; Kathy K. Niakan; et al. Frequent loss of heterozygosity in CRISPR-Cas9–edited early human embryos. Proceedings of the National Academy of Sciences 2021, 118, e2004832117, 10.1073/pnas.2004832117.
- Luis Uriel Gonzalez-Avila; Juan Manuel Vega-López; Leda Ivonne Pelcastre-Rodríguez; Omar Alejandro Cabrero-Martínez; Cecilia Hernández-Cortez; Graciela Castro-Escarpulli; The Challenge of CRISPR-Cas Toward Bioethics. Frontiers in Microbiology 2021, 12, e2004832117, 10.3389/fmicb.2021.657981.
- Samantha Elaine Noll; Philosophy Documentation Center; Review of Genetic Ethics: An Introduction, by Colin Farrelly. Essays in Philosophy 2019, 20, 245-250, 10.7710/1526-0569.1638.
- Gabriela Arguedas-Ramírez; Ethics and Global Governance of Human Germline Genome Editing: The Problem of Techno-Scientific Colonialist Paternalism. The CRISPR Journal 2020, 3, 83-88, 10.1089/crispr.2019.0045.
- George Q. Daley; Robin Lovell-Badge; Julie Steffann; After the Storm — A Responsible Path for Genome Editing. New England Journal of Medicine 2019, 380, 897-899, 10.1056/nejmp1900504.
- Should the Rich Be Allowed to Buy the Best Genes? . Airmail. Retrieved 2021-12-30
- Ritika Luthra; Simran Kaur; Kriti Bhandari; Applications of CRISPR as a potential therapeutic. Life Sciences 2021, 284, 119908, 10.1016/j.lfs.2021.119908.
- Mingtao Zhang; Emily A. Eshraghian; Omar Al Jammal; Zhibi Zhang; Xiao Zhu; CRISPR technology: The engine that drives cancer therapy. Biomedicine & Pharmacotherapy 2020, 133, 111007, 10.1016/j.biopha.2020.111007.
- Jason L. Vassy; Kurt Christensen; Erica F. Schonman; Carrie L. Blout; Jill O. Robinson; Joel B. Krier; Pamela M. Diamond; Matthew Lebo; Kalotina Machini; Danielle R. Azzariti; et al.Dmitry DukhovnyDavid W. BatesCalum A. MacraeMichael F. MurrayHeidi L. RehmAmy L. McGuireRobert C. Greenfor the MedSeq Project The Impact of Whole-Genome Sequencing on the Primary Care and Outcomes of Healthy Adult Patients. Annals of Internal Medicine 2017, 167, 159-169, 10.7326/m17-0188.
- Monti, M.; Redi, C.A. Il rene suino per gli umani interroga anche l’etica. La Lettura 2021, 519, 17 (in Italian).
- Beverley A. Townsend; Human genome editing: how to prevent rogue actors. BMC Medical Ethics 2020, 21, 1-10, 10.1186/s12910-020-00527-w.
- The Universal Declaration on Bioethics and Human Rights . UNESCO. Retrieved 2021-12-30
- CRISPR Democracy: Gene Editing and the Need for Inclusive Deliberation . Issues Sci. Technol. 2015, 32, 37.. Retrieved 2021-12-30
- Wagner, B.. Ethics as an Escape from Regulation: From Ethics-Washing to Ethics-Shopping?; Hildebrandt, M., Eds.; Amsterdam University Press: The Netherlands, 2018; pp. 84-89.




