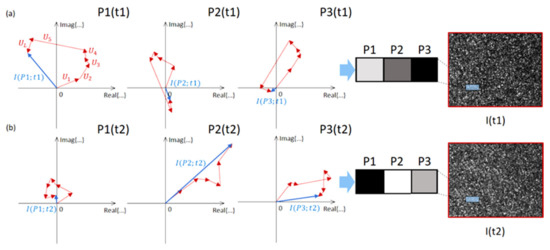
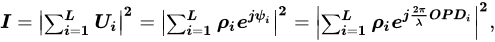The development of more sensitive methodologies, capable of quickly detecting and monitoring a microbial population present in a specific biological matrix, as well as performing to allow for the study of all its metabolic changes (e.g., during the formation of biofilm) to occur, is an essential requirement for both well-being and the food industry. Two techniques, in particular, have gained the attention of scientists: The first is “biospeckle”, an optical technique representing an innovative tool for applications in food quality, food safety, and nutraceuticals. A second technique with great chances is the “biofilm electrostatic test” (BET). BET undoubtedly represents a fast, simple, and highly reproducible tool suitable for admitting the evaluation of the in vitro bacterial capacity in order to adhere through an electrostatic interaction with a pyro-electrified carrier after only 2 h of incubation.
1. Introduction
Bacteria are present in nature and the environment. Most of the microorganisms carry out essential activities in nature, and many are closely associated with plants or animals in beneficial relations. These well-known microorganisms are the first example of the biotechnological process, concurring through the fermentation of some matrices, such as milk, meat, fish, grapevine, and cereals, in order to produce foods that are vital elements in the human diet. The use of these microorganisms is also essential for the production of new functional products or ingredients containing, for instance, immobilized probiotic/synbiotic formulae entrapped in safe matrices. Additionally, in this form, their vitality is preserved during some food technological processes or along the gastrointestinal tract.
Simultaneously, several bacteria are undesired or even pathogens, concurring in some cases to alter the quality and healthy characteristics of food, and in many instances to cause important diseases (human, animal or even vegetal). Generally, they are found in all of the environments, in the microbiota (both animal and vegetal) and the water polluted with fecal material. Microbial diseases constitute one of the primary causes of death in many developing countries of the world. These microorganisms are resistant to environmental conditions. In addition, most of the human population is entirely susceptible, and the diseases they cause are severe with a high fatality rate. A considerable amount of these lethal microorganisms could readily be grown and conserved for many years. Moreover, the number of pathogens with resistance to conventional drugs is increasing. Generally, bacteria are present in the planktonic form. However, as their number increases, the microbial cells begin to organize themselves into structured microbial communities, encased in an extracellular matrix known as biofilms, capable of colonizing every surface animal, vegetal or even mineral, sediments, soil, as well as all of the varieties of biomedical implants and transcutaneous devices
[1][2]. The physiology of microorganisms when forming the biofilm is different compared to the planktonic one. In this “new” form, they tend to be more resistant to the treatment with antibiotics and other biocides, which are protected by the envelope formed by exopolysaccharides and proteins
[3][4]. Therefore, the development and use of more sensitive methodologies, capable of quickly detecting and monitoring a microbial population present in a specific biological matrix, as well as performing to allow for the study of all its metabolic changes (e.g., in the case of the formation of biofilm) to occur, is an essential requirement for both health and the food industry. Traditionally, the bio-molecular methodologies can replace standard methods of essential microbiology. However, although these methods are certainly faster than the former, they require some preliminary steps (DNA extraction, gene amplification by PCR-RT, material analysis), which can still cause an undesired loss of time.
In the case of biofilms, conventional methods based on plating and microscopic counting are suitable for the study and detection of the number of bacteria adhering onto eukaryotic cells or inanimate surfaces.
In the literature, the best-known methods that study and evaluate the biofilm formation are based on microtiter plates, developed by Madilyn Fletcher
[5], in which the measurement of the biomass attached quantifies the biofilm
[6][7]. In the first developed procedure, bacterial cells are grown in a polystyrene microtiter plate
[8].
The planktonic cells are washed away for different observation times before staining the biomass attached to the wells′ surface to remove the non-adherent cells. The biofilm biomass can alternatively be quantified by detachment and subsequent plating. One major limitation of this technique is that the extracellular polymeric substances (EPS) can cover parts of the biomass derived from the settled cells on the bottom of the well. Therefore, this biomass could come from the biofilm-forming process, thus introducing a non-negligible error. The Calgary biofilm device (CBD)
[9], based on the application of pegs that fit into the wells of the microtiter plate, could prevent the biofilm formation from the settled cells. This system produces 96 equivalent biofilms, for the assay of antibiotic sensitivity through the standard 96-well technology. In this way, the biofilm formed on the pegs arises from a sessile development rather than cell sedimentation
[10][11][12].
Moreover, biofilm formation is primarily investigated by quantitative microbiology and scanning electron microscopy. Nevertheless, some issues remain as the recovery of bacterial cells, which takes place through sonication, usually retrieves only a part of them. Furthermore, the physiological properties of the detached population may not reflect the physiology of sessile cells since different microbial populations may present diverse adhesions and thus, various detachments of material
[13][14].
Another technology that has been developed more recently consists of the so-called Biofilm Ring Test
[15][16] (Saint-Beauzire, France), which uses the immobilization of magnetic beads by the biofilm matrix, while growing in vitro
[17][18].
It is possible to evaluate the capacity for biofilm formation through indirect methods. When the cell-matrix is not strong enough to trap and hold the microspheres, for instance, to overcome the attractive forces exerted by external magnets, this gives rise to a weak biofilm. Conversely, a biofilm formed more rapidly will quickly and effectively block the microparticles
[19]. The absence of biofilm led to the attraction of beads to the center of the bottom wells and to the formation of a central point. The presence of the biofilm formation gives rise to the immobilization of beads, and the stain disappears. However, this methodology has some limitations for extensive use in a clinical setting since it requires the additional step of treating the cells with magnetic beads, resulting in the introduction of other variables that may influence the repeatability of the tests
[20].
To the best of our knowledge, these methods are laborious and require a large amount of time. In addition, the bacteria should be viable after the releasing process. Other methods, such as those based on microscopy or colorimetric approaches, are also applied after fixation and Gram staining, but they are laborious. Currently, crystal violet (CV) staining is the most widely used method for in vitro quantification of biofilm due to its relative simplicity and sensitivity
[21]. However, it is limited by evident factors, not least in the time of the procedure (24−48 h of incubation), multiple processing steps, large standard deviations of the readouts, and weak flexibility, if we do not want use it for large-scale screening.
At the present time, two techniques, in particular, have gained the attention of scientists: The first is the so-called “biospeckle”, an optical method, which is an innovative tool for applications in food quality, food safety, and nutraceuticals. Through this technique, we can evaluate and monitor—very quickly—the presence of bacteria (or their proliferation) in a solid or liquid biological matrix. Furthermore, through the decorrelation of the biospeckle, we can quantify and optimize the storage time of the probiotic bacteria encapsulated in alginate and their survival rate in simulated gastro-intestinal conditions.
In the last decade, a new family of biological applications uses ferroelectric crystals
[22], including the fungal growth stimulated by laser interference gratings
[23], and the bacterial patterning in water-based media
[24], to cite some examples. In particular, for the first time, Rega et al. demonstrated the possibility of using the pyroelectric effect in ferroelectric materials for cell patterning
[25][26][27][28][29] and soft matter manipulation
[30][31][32].
Moreover, the same authors proposed the pyroelectric effect as a successful method for what has been called the “biofilm electrostatic test” (BET)
[27][28][29][33]. This method allows for the study and monitor of the formation of a microbial biofilm, as well as the more or less aptitude of certain microorganisms in order to form a biofilm when grown in particular conditions.
2. Biospeckle Analysis
From a mathematical viewpoint, we could consider speckle a random walk phenomenon in the complex plane
[34]. When coherent light probes the rough surfaces, speckles originate on the recording device. If the surface roughness is comparable to the probe wavelength, let us model the surface under the test as consisting of L independent scatters, i.e., L independent new point sources placed in different 3D coordinates. Light scattered from each point reaches the detector after propagating through the medium (e.g., air, PBS, water), experiencing a different optical path. Therefore, the detector receives the coherent superposition of the L complex wave front contributions in the form of an intensity pattern:
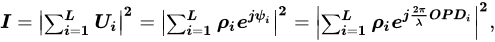
Figure 1 sketches the random walk process for the neighbor pixels of the detector, showing how much the OPD differences between each contribution govern this. Although similar gray levels could represent close-by points of a macroscopically homogeneous surface after detector quantization, the random walk process generates different results from the coherent sum. This situation is primarily due to the wavenumber factor (2π/λ) in the argument of Ui that amplifies any tiny OPD difference. Therefore, the pixels in Figure 1a are represented by different values, resulting in the typical sequence of bright and dark spots in a speckle representation.
Figure 1. Random walk in the complex plane. The process of speckle grains formation at the receiver is sketched for three close-by pixels of a homogeneous region of the image. The resultant of each coherent sum is indicated with a blue arrow. (a) t = t1. (b) t = t2.
A speckle is thought of as an impairment for imaging, and many speckle reduction techniques have been proposed over the years. However, speckle grains represent a precious source of information, which is the coherent sum sensitive to sub-micrometric time variations of the rough surface, i.e., changes occurring on the scale of the probe wavelength. In microscopy configurations, it is helpful to study the formation of speckle patterns generated when light is reflected/refracted from a scattering medium to infer information on the medium itself. Furthermore, the size, spatial, and frequency distribution of the speckle grains can be linked to the shape of the scattering object, its roughness, as well as the superficial or volumetric density
[34][35][36][37]. For these reasons, optical speckle metrology has become one of the most sensitive classes of measurement techniques, and it is adopted in many different industrial fields for non-disruptive testing (NDT) of macroscopic size objects
[38].
Among the wide variety of methods that use speckles to retrieve information on the presence of biological activity inside a specific field of view (FoV), we can distinguish between the static approaches, where speckles do not change over time, and the dynamics ones, influenced by the optical Doppler effect that leads to speckle changes over time. Therefore, the dynamic speckles contain information on the object′s movement or the motion of particles within the object.
Regarding the possibility of studying the static scattering properties of a single biological element, a sizeable numerical aperture (NA) microscopy setup is mostly required, which sacrifices the available FoV
[39][40][41][42]. However, measurements of light-scattering profiles with a scanning flow cytometer have morphologically differentiated T- and B-lymphocytes, providing the cell diameter, the ratio of nucleus to cell diameter, and the refractive index of the nucleus and cytoplasm for each cell
[41]. Furthermore, the model-specific solution of the inverse light-scattering problem (ILS)
[41] has been applied in the bacterial field. In particular, ILS retrieves the length and diameter of single
Escherichia coli bacteria relying on a hemisphere-capped cylinder model, shaping each element
[39]. The rod-like shape allowed for the classification among four different bacteria species looking at the Fourier transform light scattering (FTLS) pattern
[40]. Furthermore, a single
E. coli cell diffraction pattern has been imaged at near-atomic resolution by coherent X-rays, simply by combining each particle diffraction by X-ray free-electron lasers
[42].
Rather than looking at one single element, an ensemble characterization is required to acquire information from a large FoV in order to infer statistically relevant data. For example, in the case of bacteria, the way a cell affects light propagation has been intensely investigated
[43][44][45][46][47].
The Fresnel diffraction pattern acquired through transmission can represent a fingerprint of specific species, i.e., the captured diffraction rings are unique markers enabling classification from heterogeneous samples
[43][48]. Elastic light scattering (ELS) collects transmitted light from multiple angles to infer the size of bacteria populations (
Pseudomonas aeruginosa,
Staphylococcus aureus, and
Bacillus subtilis) of various dimensions and shapes in the absence of prior information, providing a method to monitor the environment for microbial contamination
[44].
A multispectral approach on ELS patterns from bacterial colonies has been performed
[49], although it is feasible in the absence of sample movements, and capable of providing different spectral responses via wavelength-dependent refractive indices. Similarly, a fingerprint of specific colonies can be obtained using Fourier transform Raman spectroscopy, which is fast in gaining spectra and in non-disruptive modality
[50].
Lens-less imaging has been involved in cell monitoring to achieve a large FoV, maintaining microscopic resolution, image quality, and easy implementation
[51]. However, similar to bacteria, small objects might not diffract enough light to be detected depending on the culture medium of dipping. According to this purpose, a thin wetting film lens-less imaging has been developed for micro-objects detection and counting
[52][53], such as bacteria, bragging about a considerable FoV by a compact system combined with in-line holographic reconstruction
[52].
Accuracy and specificity in classifying various populations co-existing in the same heterogeneous sample can be largely improved by widening the observable space. An excellent example in this sense is the class of methods relying on hyperspectral imaging (HIS)
[45]. HSI captures a wide range of wavelengths (from 200 nm to 2 μm) to enlarge the observable space and has been successfully applied to the static inspection of the quality of meat, fruit, and seafood, by detecting defects, imperfections, assessing ripeness, and aging, as well as the presence of bacteria colonies
[46][48][52]. These methods acquire spatial maps of the objects inside the FoV by tuning the wavelength of the light source from the ultraviolet to the NIR region and stacking the captures in the so-called hypercube. However, the large dimensionality of the hypercube makes it cumbersome to handle unless efficient data reduction is applicable (e.g., using the principal component analysis, PCA)
[52]. Furthermore, the large extent of the information stored in the hypercube can be univocally associated with a specific population of biological elements. For example, the prediction of
E. coli contamination has been determined thanks to the scattering information collected in HSI
[46]. Similarly, a NIR hyperspectral imaging system (900–1700 nm wavelength range) has been employed to predict and visualize the total Enterobacteriaceae loads in the raw chicken breast fillets
[53]. Moreover, NIR HIS is suitable for quantifying total viable counts in chicken breast fillets, leveraging the method′s rapidness and non-destructiveness characteristics
[54]. Therefore, the need for fast detection, classification, and discrimination systems for foodborne bacterial pathogens with high sensitivity and specificity is pivotal in food safety control, allowing for the HIS spread in food safety, which is deeply reviewed in
[54]. Even a multimode approach has been presented to address food safety and quality importance, the joint action of spectroscopic methodologies involving hyperspectral reflectance, fluorescence mapping, Raman, and speckle imaging to improve detection performance
[55].
3. Biofilm Electrostatic Test
The formation of bacterial biofilm and adhesive processes to several surfaces by bacteria and fungi has substantial implications in food manufacturing, as well as transformation and health-linked areas. The cellular mechanisms that regulate the formation of bacterial biofilm are still under study. In addition, their control is an objective for new specific interventions and plans to manage the problems due to the biofilm formation and presence. In particular, the food decline does not only translate into economic losses. Food safety is one of the main priorities in today′s globalized market. The interaction between three main components, bacterial cells, growth medium, and the environment surrounding the bacterial attachment surface, affects the adhesion of the bacterial cells and the subsequent biofilm. Managing the interactions between bacteria and surface allows us to have a powerful tool capable of controlling the formation of bacterial biofilm.
We recently proposed the innovative BET technique to promote rapid biofilm formation through the electrostatic interaction with charged polymer sheets. The technique induces a permanent dipole charge into thin polymer sheets by pyro-electrification (PE)
[25][26][27][28][29]. The technique appears to be versatile, reproducible, simple, and easy to use for multipurpose applications, ranging from live-cell patterning to bacteria biofilm formation tests
[25][26][27][28][29].
Figure 2 shows the schematic view of the PE process.
Figure 2. Schematic representation of the PE process steps with the corresponding charge distributions.
The process consists of slowly heating a polymer film spin-coated on a substrate of lithium niobate crystal (LN) to a temperature higher than the glass transition temperature (Tg) of the polymer. Next, the treatment of the substrate (with the film deposited on its surface) to different temperatures induces a fast temperature gradient and creates a pyro-electric macroscopic field. At this condition, the polymer molecules can reorient under the effect of the uncompensated surface pyro-electric charge of LN and, when the intermolecular motion stops, the molecules “freeze” in a poled configuration. After that, the polymer film can be peeled off from the LN substrate, producing free-standing polarized membranes. Regarding the conventional electrification techniques, the PE is electrode-free, thus simplifying the process significantly. See Ref.
[25] for more details.
The assessment of microbial capability to adhere to and form biofilms is essential for evaluating how different environmental factors may affect their vitality. Currently, the approaches used are still weak in reliability and rapidity and frequently give conflicting results. BET appears to be an easy, fast, and highly replicable instrument for estimating the in vitro capacity of bacteria in order to form biofilms through an electrostatic interaction with a pyro-electric carrier.
The rapid bacterial adhesion and formation of biofilms endorsed by BET would substantially affect health assistance, when a quick answer to an antibiogram test would allow for an immediate cure to fight those diseases giving rise to bacterial infections.
BET sheets stimulate rapid adhesion and biofilm formation by
Listeria innocua (Gram-positive) and
E. coli (Gram-negative) bacteria strain. The method admits an easy, fast, and economically efficient evaluation of biofilm formation, using the electrostatic interaction between the planktonic bacteria with the pyro-electric support
[27][29].
After the pyro-electrification process, the BET-carrier is provided by the δ + charge of the permanent dipole, which is capable of immobilizing the bacteria without any damage to their cytomembrane. The negative net charge present on the bacterial cytomembrane (COO-groups) of both Gram-positive and Gram-negative bacteria is attracted by positive polarity on each surface, shown in
Figure 3, conducting bacteria adhesion and immobilization and biofilm formation
[56]. Therefore, the bacterial adhesion and consequent biofilm formation are faster on the BET support since the positive polarization charge on the BET support increases the Coulomb interaction strength, in agreement with the DLVO theory, according to which
[57] bacteria adhere to the surface when they manage to overcome the minimum secondary energy.
Figure 3. Schematic point of view of the bacterial cells interaction model with the BET-carrier. Face (δ+) and Face (δ−) indicate the polarity of the BET-carrier faces.
The images represented in Figure 4 show that the tested BET provides a polarization field capable of immobilizing L. innocua (Figure 4a) and E. coli (Figure 4b) by promoting the formation of live biofilms already within 2 h, avoiding strenuous incubations and intermediary chemical treatments. Figure 4 shows the production of two types of Polysulfone BET sheets (thickness of about 100 μm and dimensions 2 × 2 cm2): Bare sheet (not subjected to the pyro-electrification process) representing the control; and (BET) with the positive side in contact with the bacterial suspension. The incubation of the sheet at 37 °C, at two times (2 and 4 h) and in two different Petri dishes overlaid with 500 µL of bacterial suspension, is then followed by the observation at the optical microscope and by the comparison with the control.
Figure 4. (a) Optical microscopy images of bacterial adhesion of L. innocua at two different time points on the control and BET; (b) optical microscopy images of bacterial adhesion of E. coli at two different time points on the control and BET.
The amount of adherent bacteria on the BET, consequent to the biofilm form, in both bacterial strains was visibly higher than the control at each observation time.
BET allows us to have the scenario of a 24-h biofilm of both Gram-negative and Gram-positive bacteria within 2 h and with 6-fold higher density than the control. Due to the structural integrity of the cell membrane, the BET-carrier is capable of supporting the growth of a vital biofilm even after 24 h incubation and with a growth rate 6-fold higher than the control. The live/dead staining kit admits to monitoring biofilms’ viability (see Figure 5). After 24 h of incubation, we can see that the viability of the high-density biofilms formed on the BET sheet is evident in Figure 5. The BET sheet immobilizes the planktonic bacteria more quickly than the control, favoring the biofilm formation without damaging the bacterial cell membrane even after 24 h of incubation, thus proving their biocompatibility. Conversely, chemical coatings that usually immobilize planktonic bacteria could damage their cell membrane, causing death.
Figure 5. Fluorescence microscope images of L. innocua (a) and E. coli (b) forming a biofilm on the BET sheet after 24 h incubation and live/dead staining.
Further quantitative studies evaluated the practical efficiency of the BET vector. In particular, the biomass quantity adhered on the BET vector surface was determined in the early stages of bacteria incubation. In this analysis, two bacterial suspensions of
E. coli were used with two different concentrations differing by approximately one order of magnitude in terms of optical density (OD), and the biomass amount was assessed through the CV assay
[58].
Figure 6 shows the typical optical microscope images after CV staining. The results obtained in the first 2 h demonstrate the ability of the BET vector to promote faster biofilm formation than the control surface, even in the case of highly diluted bacterial concentration
[29].
Figure 6. Crystal violet microscope images of E. coli forming a biofilm on the BET sheet vs. control after 2 h of incubation for different OD initial values. (A,B) represent the images of High-OD E.coli forming a biofilm in the control and on the BET sheet, respectively, after 2 h of incubation. (C,D) represent the images of Low-OD E.coli forming a biofilm in the control and on the BET sheet, respectively, always after 2 h of incubation.
Figure 7a,b plots the data corresponding to the biomass determination for two different OD values. Either in high OD or low OD cases, the BET-carrier effectiveness (orange columns) has been demonstrated to immobilize a greater biomass quantity for each observation time than the control, with an increase of about 30% after 2 h for both cases. Moreover, the major amount of biomass on the BET-carrier was observed for 4-h incubation times, by 60% and 40%, respectively.
Figure 7. Quantitative determination of the biofilm adhered on the sheets by the CV absorbance in the case of the control (blue columns) and BET-carrier (orange columns) at the two incubation times, in the case of high OD (a) and low OD (b). The error bars represent the standard deviation for the three replicates of the experiment.
The BET-carrier ability to produce biofilms with high biomass in a few hours, even at low bacterial concentrations, is evident, with a significant impact .