Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jiahong Lu | + 2078 word(s) | 2078 | 2021-11-19 08:01:37 | | | |
| 2 | Jessie Wu | Meta information modification | 2078 | 2021-11-30 02:43:28 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lu, J. Autophagy and Inflammatory Pathways in Macrophages. Encyclopedia. Available online: https://encyclopedia.pub/entry/16519 (accessed on 08 February 2026).
Lu J. Autophagy and Inflammatory Pathways in Macrophages. Encyclopedia. Available at: https://encyclopedia.pub/entry/16519. Accessed February 08, 2026.
Lu, Jiahong. "Autophagy and Inflammatory Pathways in Macrophages" Encyclopedia, https://encyclopedia.pub/entry/16519 (accessed February 08, 2026).
Lu, J. (2021, November 29). Autophagy and Inflammatory Pathways in Macrophages. In Encyclopedia. https://encyclopedia.pub/entry/16519
Lu, Jiahong. "Autophagy and Inflammatory Pathways in Macrophages." Encyclopedia. Web. 29 November, 2021.
Copy Citation
Autophagy as a conserved bulk degradation and recycling process, performs specific roles in macrophage to regulate innate immune response. This review focuses on the role of autophagy, both as nonselective and selective forms, in the regulation of the inflammatory and phagocytotic functions of macrophages. Specifically, the roles of autophagy in pattern recognition, cytokine release, inflammasome activation, macrophage polarization, LC3-associated phagocytosis, and xenophagy are comprehensively reviewed.
macrophage
autophagy
phagocytosis
1. Introduction
Autophagy is a highly conserved mechanism by which the cytoplasmic cargo is delivered to the lysosomes for degradation. There are at least three forms of autophagy: chaperone-mediated autophagy (CMA), microautophagy, and macroautophagy. Macroautophagy is the major autophagic degradation form that maintains the cell homeostasis and organelle quality control in eukaryotic cells. Macroautophagy (hereafter referred to as autophagy) plays crucial roles in various cellular physiological processes, including cellular metabolism, residual cargo removal, renovation in cell differentiation and development. Emerging evidence has revealed the implications of autophagy in numerous diseases, including immunological diseases, cancer, neurodegenerative diseases, cardiovascular disorders and aging [1]. One feature of autophagy is the formation of a double membrane structure called autophagosome; this formation comprises four main steps: (1). autophagosome initiation, (2) autophagosome elongation, (3) autophagosome closure, and (4) autophagosome fusion with lysosome [2]. In the initiation stage, ULK1 complex (consisting of ULK1, FAK family interacting-protein of 200 kDa (FIP200), ATG13 and ATG101) is activated and localizes in the ER [3]. Subsequently, class III phosphoinositide 3-kinase (PI3K) complex (consisting of VPS34, VPS15, Beclin 1, ATG14L and NRBF2) is activated by ULK1 to generate PI3P which recruits its binding proteins, WD repeat domain phosphoinositide-interacting protein 2 (WIPI2), and zinc-finger FYVE domain-containing protein 1 (DFCP1) [4][5]. Then two ubiquitination-like systems are activated to elongate the autophagosome. In the first system, ATG5 is covalently conjugated to ATG12, and then interacts with ATG16L1. In the second system, ubiquitin E1-like enzyme ATG7, E2-like enzyme ATG3, and ATG5–ATG12 work together to facilitate the conjugation of phosphatidylethanolamine (PE) to the autophagosome-associated protein ATG8/LC3 [6]. ATG8/LC3 changed from LC3-I to LC3-II; form by conjugating with a phosphoethanolamine during autophagy. The membrane-bound LC3-II induces the growth and closure of the autophagosome, which then fuses with lysosome via SNARE [7].
2. Autophagy and Macrophage Phagocytic Function
2.1. LC3-Associated Phagocytosis
Phagocytosis is defined as a pathway for the recognition and internalization of substrate particles (>0.5 μm) by professional phagocytes, such as macrophages or non-professional phagocytes, such as epithelium cells [8] to maintain the cell homeostasis when threatened by invaders [8]. In 2007, the Douglas R. Green Lab firstly reported that BECN1, LC3, ATG5 and ATG7 can be recruited to the phagosome in macrophage during TLR pathway activation [9]. This process has been identified as an unconventional LC3-associated phagocytosis (LAP). The characteristics of the LAP molecular requirement have been revealed. Rubicon is required for the activity of a Class III PI3K complex containing BECN1, UVRAG, and VPS34 during LAP to generate PI (3) P without ATG14 and Ambral, two conventional autophagy-related proteins. Further, ATG5, ATG3, ATG12, and ATG16L are all necessary for the conjugation of LC3 to the LAPosome [10] (Figure 1).
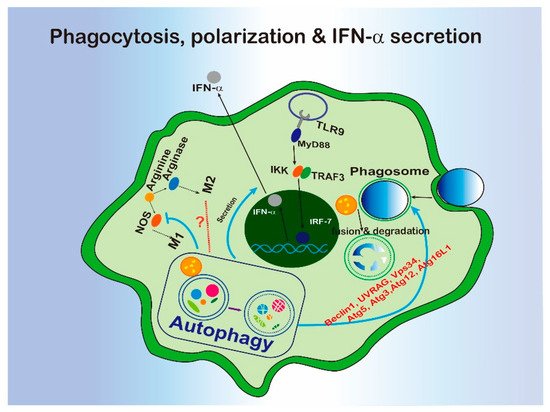
Figure 1. The interregulation of phagocytosis, macrophage polarization, or IFN-α secretion and autophagy.
Based on the biological functions of phagocytosis, LAP displays various roles in immune response (Figure 1). For example, LAP activation is associated with innate immune recognition and cellular defense system to defend against pathogens. LAP promotes antigen presentation by MHC class II molecules to T cells [11] and helps to clear pathogens via engulfment and phagosome acidification [10][9]. In terms of the inflammatory response, LAP has been implicated in DNA-induced TLR9 pathway activation. LC3 and kinase IKKα have been found to form a complex and be recruited to the endosome containing TLR9. The complex are further associated with TRAF3 and IRF7 with the help of ATG5 to promote type 1 interferon production [12]. In the tumor microenvironment, defects of LAP induced control of tumor growth by tumor-associated macrophage (TAM) through triggering pro-inflammatory gene expression and triggering a STING-mediated type I interferon response [13]. Since a series of autophagy-related proteins are involved in, LAP could be a major link between autophagy and innate immunity.
The role of LAP in dead cell clearance was reported in 2011 [14]. The study showed that LAP can be evoked by incubating macrophages with apoptotic, necrotic and RIPK3-dependent necrotic cells. The dying cells were efficiently degraded through LAP. As the defective clearance of dying cells is associated with systemic lupus erythematosus (SLE), this study found mice with LAP pathway defect displayed increased SLE-like symptoms after repeated injection of apoptotic cells [15].
2.2. Xenophagy
Autophagy was initially regarded as a non-selective process, while recent studies revealed that autophagy can also be selective. Selective autophagy is responsible for removing of specific cellular cargos, by a recognition mechanism involving autophagy receptors or adaptors [16]. According to different cargos, selective autophagy has been classified into different types, such as aggrephagy (cargos: protein aggregtes), mitophagy (cargos: mitochondria), xenophagy (cargos: pathogens), ER-phagy (cargos: endoplasmic reticulum), pexophagy (cargos: peroxisomes), and so on.
Xenophagy is a form of selective autophagy which specifically targets invading pathogens. Xenophagy in macrophage has been well characterized during Mycobacterium tuberculosis infection. Specifically, the internalized phagosomal M. tuberculosis can be released into cytosol by damaging the phagosomal membrane [80]. Escaped bacterial DNA was recognized by cytosolic sensor cGAS, which induced xenophagy via activating ubiquitination by ubiquitin ligases Parkin and Smurf1 and recruiting autophagy receptors or adaptors, such as P62 and NDP52 [122,123,124]. Not only the bacterial but also damaged phagosomes containing bacteria can be targeted by the host glycan on the phagosomal lumen [125,126]. Ultimately, bacteria were sent to the lysosome via the autophagosome for degradation. In this process, autophagy also facilitated the host to kill bacteria via generating and delivering antimicrobial peptides to the compartment [127,128]. In addition, NOD1 and NOD2 sensed invasive bacterial and induced xenophagy by recruiting ATG16L1 to the site of bacterial entry [49]. The adaptors or receptors for xenophagy are not limited to P62 and NDP52, neighbors of the BRCA1 gene 1 (NBR1) and optineurin can also serve the xenophagy process [129].
Even though xenophagy requires most of the molecular machinery involved in classical autophagy, increased susceptibility to M. tuberculosis infection was only observed in the mice with atg5 deficiency in monocyte-derived cells and neutrophils, not those lacking beclin-1 or atg14 [17][18][19]. In some cases, pathogens are able to develop various strategies to destroy autophagy for survival. For example S. flexneri can escape from xenophagy by secreting IcsB, which can inhibit bacterial recruitment to the phagophore via binding competitively to the surface protein VirG [20]. L. monocytogenes inhibits xenophagy via recruiting the ARP2/3 complex and Ena/VASP to the bacterial surface, where it masks the cell surface by binding to the cytoplasmic major vault protein (MVP) or blocking the lipidation of LC3 [21][22][23]. Salmonnella typhimurium secrets more than 30 effector proteins, resulting in the mTOR activation [24], deubiquitination of aggregates [25], and disrupting RAB1-A signal [26], and ultimately, autophagy suppression. In their most recent study, Shao Feng’s lab found that a novel protein, SopF, generated by Salmonella inhibit xenophagy by targeting the GLN124 of ATP6V0C in the V-ATPase, resulting in disruption of V-ATPase-ATG16L1 axis in xenophagy initiation [27]. So, xenophagy partially facilitates macrophage phagocytosis to clear invasive pathogens, especially when the phagosome has been damaged (Figure 2). However, xenophagy can also be blocked by flexible escape strategies developed by invaders.
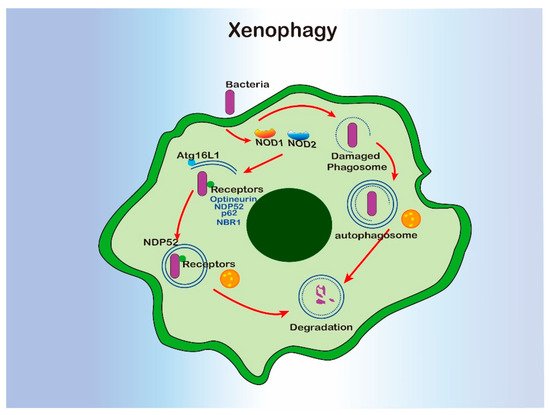
Figure 2. The interregulation of xenophagy and macrophage.
2.3. Autophagy Receptors in Macrophage
Autophagy receptors are adaptor proteins which recognize cargos and bind to LC3/GABARAP on autophagosome [16]. The selectivity of cargos is mainly achieved by the specificity of autophagy receptors in recognizing distinct cargos. As mentioned above, P62 is a key autophagy receptor involved in the inflammasome pathway regulation. In addition, P62 and other receptors or adaptors also regulate macrophage functions via modulating selective autophagy process.
Selective receptors are involved in the innate inflammation pathway. According to the multiplex proteomic profiling, P62 and TAX1BP1 were identified to be the autophagy receptors that mediated the turnover of innate adaptor TRIF and its downstream signaling in atg16l1 deficient macrophages [28]. Knockdown of tax1bp1 increased cytokines release, such as IFN-β and IL-1β [28]. TRIM20 and TRIM21 are subsets of tripartite motif (TRIM) proteins, and they also acted as autophagic receptors to recognize inflammasome components or dimeric form of IRF3, delivering them for autophagic degradation [29]. As above mentioned, CGAS could also be degraded via autophagy after being recognized by P62. Conversely, P62-mediated autophagic degradation of CGAS enhanced the activation of type interferon I signaling [30]. Under LPS-induced inflammation, P62 interacts with iNOS and facilitates autophagic degradation of iNOS [31]. In cases of inflammasome pathway activation, P62 usually acted as an important receptor for inflammasome or related proteins degradation and so that to regulate inflammation response [32][33]. Inflammasome pathway can be easily activated by released mitochondrial DNA, and P62 has been reported to be involved in the inflammasome modulation as a receptor of mitophagy. P62-dependent mitophagy dysfunction caused inflammasome-induced IL-1β-dependent inflammation [34].
Several selective receptors are involved in xenophagy to mediate innate immunity response in macrophage. P62 and NDP52 are two classic receptors implicated in xenophagy. P62 assembles on the microbes as receptors once the cells were affected by pathogens, such as Salmonella [35] and Mycobacteria [36]. NDP52 can be recruited as xenophagy receptors for Salmonella [37]. NDP52 has been proved to recruit ULK complex to the cytosol-invading bacteria and initiate autophagy [38]. Optineurin (OPTN) is another autophagy receptor that is implicated in xenophagy and controlling of TNF, NF-κB, and IFN signaling in macrophage [39][40][41]. TBK1 regulates OPTN phosphorylation to recognize cytosolic Salmonella enterica and trigger xenophagy [42][43]. During innate immunity response, OPTN served as a negatively regulator of NF-κB by promoting the xenophagy [44][45]. Another autophagy receptor, NBR1, was found to bind with viral capsid protein and particles of cauliflower mosaic virus (CaMV) for autophagic degradation [46]. TRIM5, a well-known retroviral restriction factor, was proposed to be a selective autophagy receptor targeting HIV-1 capsids for autophagic degradation [47]. Recently, V-ATPase was identified as the sensor of invading pathogen to recruit ATG16L to initiate xenophagy [27].
2.4 Autophagy and Macrophagic Metabolism
Emerging studies have revealed the crucial role of metabolic reprogramming in macrophage activation, which is known as immunometabolism [48]. For example, in amino acid metabolism, arginine is converted to NO by iNOS in M1 macrophage, but metabolized by arginase-1 in M2 macrophage [49][50]. M1 macrophage shows enhanced glycolytic metabolism and impaired mitochondrial oxidative phosphorylation (OXPHOS) [51][52]. In addition, the ATP produced in M1 cells via glycolytic metabolism feeds the pentose phosphate pathway (PPP) [53]. Fatty acid synthesis (FAS) organizes the plasma membranes under different inflammatory responses to regulate the inflammatory signaling, adhesion, and migration of macrophages [54]. Fatty acid oxidation (FAO) is needed for NLRP3 inflammasome activation [54]. These studies highlight the view that macrophagic metabolism status is tightly associated with macrophage functions.
Autophagy is an important part of cellular metabolism system that can be activated to supply materials and energy during nutrient deficiency. Therefore, the relationship between autophagy, metabolism, and macrophage function is a hotspot being noticed and investigated. In 2018, the “Autophagy, Inflammation, and Metabolism” (AIM) Center at the University of New Mexico has been established to specifically study this topic (ref: Autophagy, Inflammation, and Metabolism (AIM) Center of Biomedical Research Excellence). Further, mTOR functions as a key homeostatic regulator in nutrient signals and metabolic processes for cell growth [55], and its inhibition induce autophagy. As mentioned in the previous part, the mTOR pathway also regulates macrophage inflammatory pathway and polarization, which could be a possible link between cellular metabolism and macrophage function. AMP-activated protein kinase (AMPK) is a main sensor of cellular energy status and maintain metabolic balance [56]. AMPK can control cell growth by inhibiting mTOR pathway via phosphorylation of TSC2 or RAPTOR [57][58]. Therefore, AMPK can be involved into autophagy indirectly through acting on mTOR pathway. In addition, AMPK can also phosphorylate ULK1 complex directly to activate autophagy [59]. According to previous studies, AMPK is regarded as a suppressor of inflammation in immune cells including macrophage [60], indicating a potential connection among autophagy, cellular metabolism and macrophage inflammation. Besides, growing evidence has revealed that autophagy can regulate immune cell differentiation by alteration of metabolic states in immune cells [61]. Collectively, upon mTOR activation, immune cells show decreased autophagy, but increased cellular glycolytic and pro-inflammatory response. However, AMPK induction induce autophagy, increase cellular OXPHOS and anti-inflammatory response [61]. Moreover, increased lipophagy (autophagic target on lipid specifically) has been proved to accelerate macrophage cholesterol efflux so that to reduce macrophage foam cells in atherosclerosis [62]. Although lots of evidence is being uncovered, the regulation network between autophagy, metabolism, and macrophage function has not been well understood. The key metabolic products that link macrophage function and autophagy would be an interesting topic to explore.
References
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527.
- Mintern, J.; Villadangos, J.A. Autophagy and mechanisms of effective immunity. Front. Immunol. 2012, 3, 60.
- Karanasios, E.; Walker, S.A.; Okkenhaug, H.; Manifava, M.; Hummel, E.; Zimmermann, H.; Ahmed, Q.; Domart, M.-C.; Collinson, L.; Ktistakis, N.T. Autophagy initiation by ulk complex assembly on er tubulovesicular regions marked by atg9 vesicles. Nat. Commun. 2016, 7, 12420.
- Mizushima, N.; Noda, T.; Yoshimori, T.; Tanaka, Y.; Ishii, T.; George, M.D.; Klionsky, D.J.; Ohsumi, M.; Ohsumi, Y. A protein conjugation system essential for autophagy. Nature 1998, 395, 395.
- Mizushima, N.; Kuma, A.; Kobayashi, Y.; Yamamoto, A.; Matsubae, M.; Takao, T.; Natsume, T.; Ohsumi, Y.; Yoshimori, T. Mouse apg16l, a novel wd-repeat protein, targets to the autophagic isolation membrane with the apg12-apg5 conjugate. J. Cell Sci. 2003, 116, 1679–1688.
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The atg12-atg5 conjugate has a novel e3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007, 282, 37298–37302.
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178.
- Gordon, S. Phagocytosis: An immunobiologic process. Immunity 2016, 44, 463–475.
- Sanjuan, M.A.; Dillon, C.P.; Tait, S.W.; Moshiach, S.; Dorsey, F.; Connell, S.; Komatsu, M.; Tanaka, K.; Cleveland, J.L.; Withoff, S. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007, 450, 1253.
- Martinez, J.; Malireddi, R.S.; Lu, Q.; Cunha, L.D.; Pelletier, S.; Gingras, S.; Orchard, R.; Guan, J.-L.; Tan, H.; Peng, J. Molecular characterization of lc3-associated phagocytosis reveals distinct roles for rubicon, nox2 and autophagy proteins. Nat. Cell Biol. 2015, 17, 893.
- Romao, S.; Gasser, N.; Becker, A.C.; Guhl, B.; Bajagic, M.; Vanoaica, D.; Ziegler, U.; Roesler, J.; Dengjel, J.; Reichenbach, J. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged mhc ii antigen processing. J. Cell Biol. 2013, 203, 757–766.
- Hayashi, K.; Taura, M.; Iwasaki, A. The interaction between ikkα and lc3 promotes type i interferon production through the tlr9-containing laposome. Sci. Signal. 2018, 11, eaan4144.
- Cunha, L.D.; Yang, M.; Carter, R.; Guy, C.; Harris, L.; Crawford, J.C.; Quarato, G.; Boada-Romero, E.; Kalkavan, H.; Johnson, M.D. Lc3-associated phagocytosis in myeloid cells promotes tumor immune tolerance. Cell 2018, 175, 429–441.
- Martinez, J.; Almendinger, J.; Oberst, A.; Ness, R.; Dillon, C.P.; Fitzgerald, P.; Hengartner, M.O.; Green, D.R. Microtubule-associated protein 1 light chain 3 alpha (lc3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA 2011, 108, 17396–17401.
- Martinez, J.; Cunha, L.D.; Park, S.; Yang, M.; Lu, Q.; Orchard, R.; Li, Q.-Z.; Yan, M.; Janke, L.; Guy, C. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 2016, 533, 115.
- Stolz, A.; Ernst, A.; Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014, 16, 495–501.
- Watson, R.O.; Manzanillo, P.S.; Cox, J.S. Extracellular m. Tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012, 150, 803–815.
- Watson, R.O.; Bell, S.L.; MacDuff, D.A.; Kimmey, J.M.; Diner, E.J.; Olivas, J.; Vance, R.E.; Stallings, C.L.; Virgin, H.W.; Cox, J.S. The cytosolic sensor CGAS detects mycobacterium tuberculosis DNA to induce type i interferons and activate autophagy. Cell Host Microbe 2015, 17, 811–819.
- Manzanillo, P.S.; Ayres, J.S.; Watson, R.O.; Collins, A.C.; Souza, G.; Rae, C.S.; Schneider, D.S.; Nakamura, K.; Shiloh, M.U.; Cox, J.S. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013, 501, 512.
- Franco, L.H.; Nair, V.R.; Scharn, C.R.; Xavier, R.J.; Torrealba, J.R.; Shiloh, M.U.; Levine, B. The ubiquitin ligase smurf1 functions in selective autophagy of mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe 2017, 21, 59–72.
- Gomes, L.C.; Dikic, I. Autophagy in antimicrobial immunity. Mol. Cell 2014, 54, 224–233.
- Chauhan, S.; Kumar, S.; Jain, A.; Ponpuak, M.; Mudd, M.H.; Kimura, T.; Choi, S.W.; Peters, R.; Mandell, M.; Bruun, J.-A. Trims and galectins globally cooperate and trim16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev. Cell 2016, 39, 13–27.
- Ponpuak, M.; Davis, A.S.; Roberts, E.A.; Delgado, M.A.; Dinkins, C.; Zhao, Z.; Virgin IV, H.W.; Kyei, G.B.; Johansen, T.; Vergne, I. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 2010, 32, 329–341.
- Alonso, S.; Pethe, K.; Russell, D.G.; Purdy, G.E. Lysosomal killing of mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc. Natl. Acad. Sci. USA 2007, 104, 6031–6036.
- Travassos, L.H.; Carneiro, L.A.; Ramjeet, M.; Hussey, S.; Kim, Y.-G.; Magalhães, J.G.; Yuan, L.; Soares, F.; Chea, E.; Le Bourhis, L. Nod1 and nod2 direct autophagy by recruiting atg16l1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010, 11, 55.
- Ma, Y.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Autophagy and cellular immune responses. Immunity 2013, 39, 211–227.
- Castillo, E.F.; Dekonenko, A.; Arko-Mensah, J.; Mandell, M.A.; Dupont, N.; Jiang, S.; Delgado-Vargas, M.; Timmins, G.S.; Bhattacharya, D.; Yang, H. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc. Natl. Acad. Sci. USA 2012, 109, E3168–E3176.
- Kimmey, J.M.; Huynh, J.P.; Weiss, L.A.; Park, S.; Kambal, A.; Debnath, J.; Virgin, H.W.; Stallings, C.L. Unique role for atg5 in neutrophil-mediated immunopathology during m. Tuberculosis infection. Nature 2015, 528, 565.
- Ogawa, M.; Yoshimori, T.; Suzuki, T.; Sagara, H.; Mizushima, N.; Sasakawa, C. Escape of intracellular shigella from autophagy. Science 2005, 307, 727–731.
- Yoshikawa, Y.; Ogawa, M.; Hain, T.; Yoshida, M.; Fukumatsu, M.; Kim, M.; Mimuro, H.; Nakagawa, I.; Yanagawa, T.; Ishii, T.; et al. Listeria monocytogenes acta-mediated escape from autophagic recognition. Nat. Cell Biol. 2009, 11, 1233–1240.
- Dortet, L.; Mostowy, S.; Louaka, A.S.; Gouin, E.; Nahori, M.-A.; Wiemer, E.A.; Dussurget, O.; Cossart, P. Recruitment of the major vault protein by inlk: A listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 2011, 7, e1002168.
- Mitchell, G.; Ge, L.; Huang, Q.; Chen, C.; Kianian, S.; Roberts, M.F.; Schekman, R.; Portnoy, D.A. Avoidance of autophagy mediated by plca or acta is required for listeria monocytogenes growth in macrophages. Infect. Immun. 2015, 83, 2175–2184.
- Casanova, J.E. Bacterial autophagy: Offense and defense at the host–pathogen interface. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 237–243.
- Mesquita, F.S.; Thomas, M.; Sachse, M.; Santos, A.J.; Figueira, R.; Holden, D.W. The salmonella deubiquitinase ssel inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 2012, 8, e1002743.
- Feng, Z.-Z.; Jiang, A.-J.; Mao, A.-W.; Feng, Y.; Wang, W.; Li, J.; Zhang, X.; Xing, K.; Peng, X. The salmonella effectors ssef and sseg inhibit rab1a-mediated autophagy to facilitate intracellular bacterial survival and replication. J. Biol. Chem. 2018, 293, 9662–9673.
- Xu, Y.; Zhou, P.; Cheng, S.; Lu, Q.; Nowak, K.; Hopp, A.-K.; Li, L.; Shi, X.; Zhou, Z.; Gao, W.; et al. A bacterial effector reveals the v-atpase-atg16l1 axis that initiates xenophagy. Cell 2019, 178, 552–566.
- Samie, M.; Lim, J.; Verschueren, E.; Baughman, J.M.; Murthy, A. Selective autophagy of the adaptor trif regulates innate inflammatory signaling. Nat. Immunol. 2018, 19, 246–254.
- Kimura, T.; Jain, A.; Choi, S.W.; Mandell, M.A.; Johansen, T.; Deretic, V. Trim-directed selective autophagy regulates immune activation. Autophagy 2017, 13, 989–990.
- Chen, M.; Meng, Q.; Qin, Y.; Liang, P.; Tan, P.; He, L.; Zhou, Y.; Chen, Y.; Huang, J.; Wang, R.-F. Trim14 inhibits cgas degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol. Cell 2016, 64, 105–119.
- Wang, J.; Wu, M.-Y.; Su, H.; Lu, J.; Chen, X.; Tan, J.; Lu, J.-H. Inos interacts with autophagy receptor p62 and is degraded by autophagy in macrophages. Cells 2019, 8, 1255.
- Shi, C.-S.; Shenderov, K.; Huang, N.-N.; Kabat, J.; Abu-Asab, M.; Fitzgerald, K.A.; Sher, A.; Kehrl, J.H. Activation of autophagy by inflammatory signals limits il-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 2012, 13, 255.
- Liu, T.; Tang, Q.; Liu, K.; Xie, W.; Liu, X.; Wang, H.; Wang, R.-F.; Cui, J. Trim11 suppresses aim2 inflammasome by degrading aim2 via p62-dependent selective autophagy. Cell Rep. 2016, 16, 1988–2002.
- Zhong, Z.; Umemura, A.; Sanchez-Lopez, E.; Liang, S.; Shalapour, S.; Wong, J.; He, F.; Boassa, D.; Perkins, G.; Ali, S.R. Nf-κb restricts inflammasome activation via elimination of damaged mitochondria. Cell 2016, 164, 896–910.
- Ishimura, R.; Tanaka, K.; Komatsu, M. Dissection of the role of p62/sqstm1 in activation of nrf2 during xenophagy. FEBS Lett. 2014, 588, 822–828.
- Chai, Q.; Wang, X.; Qiang, L.; Zhang, Y.; Ge, P.; Lu, Z.; Zhong, Y.; Li, B.; Wang, J.; Zhang, L.; et al. A mycobacterium tuberculosis surface protein recruits ubiquitin to trigger host xenophagy. Nat. Commun 2019, 10, 1973.
- Verlhac, P.; Gregoire, I.P.; Azocar, O.; Petkova, D.S.; Baguet, J.; Viret, C.; Faure, M. Autophagy receptor ndp52 regulates pathogen-containing autophagosome maturation. Cell Host Microbe 2015, 17, 515–525.
- Ravenhill, B.J.; Boyle, K.B.; von Muhlinen, N.; Ellison, C.J.; Masson, G.R.; Otten, E.G.; Foeglein, A.; Williams, R.; Randow, F. The cargo receptor ndp52 initiates selective autophagy by recruiting the ulk complex to cytosol-invading bacteria. Mol. Cell 2019, 74, 320–329.
- Netra, P.M.; Guozhi, Z.; Paul, R.M.; Maria Letizia, G.T.; Marie, P.; Damien, A.; Jonathan, D.A.; Ivana, M. The tbk1-binding domain of optineurin promotes type i interferon responses. FEBS Lett. 2016, 590, 1498–1508.
- Slowicka, K.; Vereecke, L.; van Loo, G. Cellular functions of optineurin in health and disease. Trends Immunol. 2016, 37, 621–633.
- Zhu, G.; Wu, C.J.; Zhao, Y.; Ashwell, J.D. Optineurin negatively regulates tnfalpha- induced nf-kappab activation by competing with nemo for ubiquitinated rip. Curr. Biol. 2007, 17, 1438–1443.
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts salmonella growth. Science 2011, 333, 228–233.
- Pilli, M.; Arko-Mensah, J.; Ponpuak, M.; Roberts, E.; Master, S.; Mandell, M.A.; Dupont, N.; Ornatowski, W.; Jiang, S.; Bradfute, S.B.; et al. Tbk-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012, 37, 223–234.
- Cao, Z.; Xiong, J.; Takeuchi, M.; Kurama, T.; Goeddel, D.V. Traf6 is a signal transducer for interleukin-1. Nature 1996, 383, 443–446.
- Gottipati, S.; Rao, N.L.; Fung-Leung, W.P. Irak1: A critical signaling mediator of innate immunity. Cell Signal. 2008, 20, 269–276.
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by nbr1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035.
- Mandell, M.A.; Kimura, T.; Jain, A.; Johansen, T.; Deretic, V. Trim proteins regulate autophagy: Trim5 is a selective autophagy receptor mediating hiv-1 restriction. Autophagy 2014, 10, 2387–2388.
- Van den Bossche, J.; O’Neill, L.A.; Menon, D. Macrophage immunometabolism: Where are we (going)? Trends Immunol. 2017, 38, 395–406.
- Arts, R.J.; Novakovic, B.; Ter Horst, R.; Carvalho, A.; Bekkering, S.; Lachmandas, E.; Rodrigues, F.; Silvestre, R.; Cheng, S.C.; Wang, S.Y.; et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 2016, 24, 807–819.
- Van den Bossche, J.; Lamers, W.H.; Koehler, E.S.; Geuns, J.M.; Alhonen, L.; Uimari, A.; Pirnes-Karhu, S.; Van Overmeire, E.; Morias, Y.; Brys, L.; et al. Pivotal advance: Arginase-1-independent polyamine production stimulates the expression of il-4-induced alternatively activated macrophage markers while inhibiting lps-induced expression of inflammatory genes. J. Leukoc. Biol. 2012, 91, 685–699.
- Lampropoulou, V.; Sergushichev, A.; Bambouskova, M.; Nair, S.; Vincent, E.E.; Loginicheva, E.; Cervantes-Barragan, L.; Ma, X.; Huang, S.C.; Griss, T.; et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 2016, 24, 158–166.
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces il-1beta through hif-1alpha. Nature 2013, 496, 238–242.
- Haschemi, A.; Kosma, P.; Gille, L.; Evans, C.R.; Burant, C.F.; Starkl, P.; Knapp, B.; Haas, R.; Schmid, J.A.; Jandl, C.; et al. The sedoheptulose kinase carkl directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012, 15, 813–826.
- Wei, X.; Song, H.; Yin, L.; Rizzo, M.G.; Sidhu, R.; Covey, D.F.; Ory, D.S.; Semenkovich, C.F. Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature 2016, 539, 294–298.
- Ben-Sahra, I.; Manning, B.D. Mtorc1 signaling and the metabolic control of cell growth. Curr. Opin. Cell Biol. 2017, 45, 72–82.
- Garcia, D.; Shaw, R.J. Ampk: Mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell 2017, 66, 789–800.
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. Ampk phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226.
- Zang, M.; Xu, S.; Maitland-Toolan, K.A.; Zuccollo, A.; Hou, X.; Jiang, B.; Wierzbicki, M.; Verbeuren, T.J.; Cohen, R.A. Polyphenols stimulate amp-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic ldl receptor-deficient mice. Diabetes 2006, 55, 2180–2191.
- Hardie, D.G. Ampk and autophagy get connected. EMBO J. 2011, 30, 634–635.
- O’Neill, L.A.; Hardie, D.G. Metabolism of inflammation limited by ampk and pseudo-starvation. Nature 2013, 493, 346–355.
- Riffelmacher, T.; Richter, F.C.; Simon, A.K. Autophagy dictates metabolism and differentiation of inflammatory immune cells. Autophagy 2018, 14, 199–206.
- Narabayashi, K.; Ito, Y.; Eid, N.; Maemura, K.; Inoue, T.; Takeuchi, T.; Otsuki, Y.; Higuchi, K. Indomethacin suppresses lamp-2 expression and induces lipophagy and lipoapoptosis in rat enterocytes via the er stress pathway. J. Gastroenterol. 2015, 50, 541–554.
More
Information
Subjects:
Cell Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
30 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




