Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alexander Pekarsky | + 1204 word(s) | 1204 | 2021-11-15 05:17:24 | | | |
| 2 | Bruce Ren | Meta information modification | 1204 | 2021-11-23 01:34:23 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Pekarsky, A. Potential Functionality of the MagR Protein. Encyclopedia. Available online: https://encyclopedia.pub/entry/16269 (accessed on 07 February 2026).
Pekarsky A. Potential Functionality of the MagR Protein. Encyclopedia. Available at: https://encyclopedia.pub/entry/16269. Accessed February 07, 2026.
Pekarsky, Alexander. "Potential Functionality of the MagR Protein" Encyclopedia, https://encyclopedia.pub/entry/16269 (accessed February 07, 2026).
Pekarsky, A. (2021, November 22). Potential Functionality of the MagR Protein. In Encyclopedia. https://encyclopedia.pub/entry/16269
Pekarsky, Alexander. "Potential Functionality of the MagR Protein." Encyclopedia. Web. 22 November, 2021.
Copy Citation
Recent findings have sparked great interest in the putative magnetic receptor protein MagR. However, in vivo experiments have revealed no magnetic moment of MagR at room temperature. Nevertheless, the interaction of MagR and MagR fusion proteins with silica-coated magnetite beads have proven useful for protein purification.
magnetic receptor protein (MagR)
Escherichia coli
magnetism
affinity chromatography
SQUID
1. Introduction
Iron–sulfur (Fe–S) cluster proteins are important for numerous physiological processes and are present in most known prokaryotic and eukaryotic cells [1][2][3]. The iron atoms in [2Fe–2S] clusters have been reported to interact through antiferromagnetic coupling [4]. Only recently, the Fe–S cluster protein MagR (magnetic receptor) came into spotlight [5]. The authors proposed a possible answer to the question on navigation of migratory animals. They reported that MagR, a small (~14 kDa) [2Fe–2S] protein from pigeons with homologs in numerous species, forms a ferrimagnetic, multimeric complex that responds to magnetic fields in vitro. Qin et al. also showed that the MagR protein and a MagR/Cryptochrome complex can be isolated and enriched from a complex matrix by silica-coated magnetite (SiO2–Fe3O4) beads [5]. Later, MagR fusion proteins were successfully captured from a complex matrix [6][7].
Since its discovery, the physical capabilities of MagR have been intensively questioned. When MagR constructs were subjected to magnetic stimuli in mammalian cells, they were not able to induce significant membrane channel activity in a magnetic field [8], in contrast to previous results [9]. The biologist Markus Meister considered the plausibility of magnetic sensing of MagR by calculations based on simple physical principles [10]. He found the number of iron atoms in the postulated assembly of MagR proteins [5] to be too low to even sense magnetic fields sufficiently [10]. Then, Winklhofer and Mouritsen argued that the weak exchange interactions among [2Fe–2S] clusters of adjacent proteins may only lead to spontaneous magnetization only below a few Kelvin, but not around room temperature [11]. Interestingly, one recent theory states that radical pairs might enable sensing of magnetic fields through induction of magnetic fluctuation in the MagR structure rather than permanent magnetism [12].
Until now, the magnetic behavior of MagR has not been tested at low temperatures, which could give clearer indications on a potential magnetic behavior. Additionally, the stated usability of MagR fusion proteins for protein capture with magnetic beads [6][7] requires further characterization and comparison to state-of-the-art affinity downstream processing methods to reveal potential drawbacks or benefits.
2. Evaluation of MagR Capture from a Complex Matrix
Overexpression of hexa-histidine-tagged (his-tag) dMagR and clMagR in E. coli was clearly visible with bands around 14 kDa in SDS-PAGE analysis (Figure 1a). Despite codon optimization, clMagR-his was mainly produced as insoluble inclusion bodies and could not be further investigated (Figure 1a). Binding studies with dMagR-his on SiO2-Fe3O4 beads showed that the protein was enriched from E. coli lysates. However, many host-cell proteins also adsorbed nonspecifically to the beads (Figure 1a). When we compared the efficiency of the magnetic bead capture with a state-of-the-art IMAC capture, we found that the IMAC capture was much more specific, and SDS-PAGE indicated a product with higher purity (Figure 1b). High absorption of dMagR-his at 320 nm clearly indicated the presence of Fe–S clusters in the protein. Binding studies with dMagR without his-tag underlined that protein binding occurred also without his-tag on beads, but again with many host-cell protein impurities. To shed more light on the binding conditions of MagR on beads, we performed binding studies with IMAC-purified dMagR-his in different buffers. We found that dMagR-his bound to magnetic beads between pH 5–11 in the presence of up to 2 M NaCl or 1 M (NH4)2SO4. Binding was only hindered at pH 12. Based on these results, we hypothesize very strong ionic interactions to be the reason for MagR binding, rather than specific magnetic interactions.
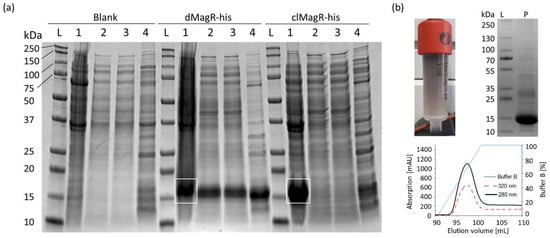
Figure 1. Evaluation of MagR purification from a complex matrix. (a) SDS-PAGE analysis of magnetic bead purification of Blank, dMagR-his and clMagR-his from cell disruption supernatant. White rectangles show respective target proteins in the applied cell pellet. Equivalent volumes of sample were applied for each respective lane (1–4). The following samples per lane are seen for each target sample: lane L: protein ladder; lane 1 (3 µL): solubilized cell pellet; lane 2 (10 µL): cell-free supernatant after cell disruption; lane 3 (10 µL): supernatant after magnetite bead precipitation; lane 4 (6 µL): bead-precipitated proteins after washing of beads. (b) Purification of dMagR-his and clMagR-his by IMAC. Coloration of IMAC column of dMagR-his is shown together with IMAC elution profile and the respective SDS-PAGE analysis of the elution pool. SDS-PAGE shows the standard protein ladder in lane L and 10 µL of the respective IMAC elution pool in lane P. Elution profiles of IMAC show absorption at 280 nm (black line), absorption at 320 nm for iron–sulfur cluster proteins (red dashed line) and relative concentration of elution buffer B (blue line).
3. Potential of MagR to Magnetize Bacterial Cells
For magnetization studies, we overexpressed the Fe–S protein dMagR without his-tag to approximately 17% of total soluble protein in E. coli (Figure 2a and Figure S3). This high intracellular content was also visible as a black–brown coloration of BL21-dMagR cell biomass and its supernatant after cell disruption (Figure 2b). Quantification by SDS-PAGE densitometry (non-MagR impurities at around 14 kDa were excluded based on a respective negative control) yielded an approximate intracellular, soluble dMagR concentration of 54 mg g−1 dry cell weight (DCW) or 5.12 pg cell−1 (1 cell~9.5·× 10−13 g DCW [13]) equivalent to 2.20·× 106 dMagR molecules cell−1. However, placing a strong neodymium magnet (50 × 50 × 12.5 mm) near the BL21-dMagR biomass suspension at room temperature resulted in no observable movement of cells towards the magnet.
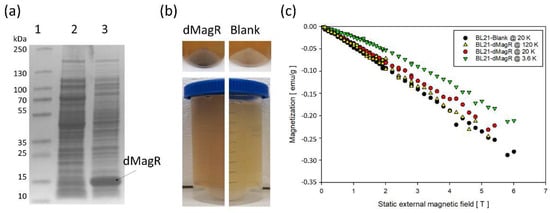
Figure 2. Potential of dMagR to magnetize bacterial cells. (a) SDS-PAGE of cell-free supernatants after cell disruption, with a clear band around 14 kDa for dMagR in Lane 3. Lane 1: Ladder; Lane 2: Supernatant of BL21-Blank; Lane 3: Supernatant of BL21-dMagR. (b) Upper part compares biomass coloration of BL21-dMagR and BL21-Blank cells. Lower part compares coloration of cell free supernatant after cell disruption. BL21-dMagR biomass and cell free supernatant show clear brownish coloration due to dMagR. (c) Isothermal, magnetic field-dependent SQUID magnetometry measurements of lyophilized BL21-Blank and BL21-dMagR at 3.6, 20 and 120 K. Conversion of emu g−1 = Am2 kg−1.
Researchers further analyzed magnetization behavior with lyophilized cells by superconducting quantum interference device (SQUID) magnetometry. Based on the vague knowledge about MagR and its applicability in cells to interact with magnetic fields at ambient conditions [8][9], we hypothesized that measurements at low temperatures of only 3.6, 20 and 120 K would give a clearer indication on a potential applicability in cells. That is due to the known temperature-dependent magnetic susceptibility of magnetic materials. The field-dependent isothermal magnetization measurements revealed a dominant diamagnetic response of BL21-Blank and BL21-dMagR cells in a static external magnetic field (emu/g = electromagnetic unit per gram DCW; emu = 10−3 Am2; emu g−1 = Am2 kg−1) (Figure 2c). The comparison of 20 K isothermal magnetization data of BL21-dMagR with corresponding BL21-Blank data revealed a rather small additional paramagnetic contribution of the former, which likely results from dMagR-bound iron in BL21-dMagR cells. As expected, this paramagnetic contribution increases with decreasing temperature, as found for BL21-dMagR cells at 3.6 K (Figure 2c). However, our results clearly show that overexpression of intracellular dMagR does not exhibit a sufficiently strong magnetic contribution to overcome the diamagnetic character of the E. coli cell, even at only 3.6 K.
References
- Konhauser, K.O.; Kappler, A.; Roden, E.E. IRON IN MICROBIAL METABOLISMS. Elements 2011, 7, 89–93.
- Lill, R.; Hoffmann, B.; Molik, S.; Pierik, A.J.; Rietzschel, N.; Stehling, O.; Uzarska, M.A.; Webert, H.; Wilbrecht, C.; Mühlenhoff, U. The role of mitochondria in cellular iron–sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1823, 1491–1508.
- Paul, V.D.; Lill, R. Biogenesis of cytosolic and nuclear iron–sulfur proteins and their role in genome stability. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1853, 1528–1539.
- Pandelia, M.-E.; Lanz, N.; Booker, S.J.; Krebs, C. Mössbauer spectroscopy of Fe/S proteins. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1853, 1395–1405.
- Qin, S.; Yin, H.; Yang, C.; Dou, Y.; Liu, Z.; Zhang, P.; Yu, H.; Huang, Y.; Feng, J.; Hao, J.; et al. A magnetic protein biocompass. Nat. Mater. 2016, 15, 217–226.
- Jiang, M.; Zhang, L.; Wang, F.; Zhang, J.; Liu, G.; Gao, B.; Wei, D. Novel Application of Magnetic Protein: Convenient One-Step Purification and Immobilization of Proteins. Sci. Rep. 2017, 7, 13329.
- Wang, L.; Xu, H.; Liu, Z.; Sun, T.; Yuan, C.; Yang, Y.; Guo, J.; Xie, H. Magnetic immobilization of a quorum sensing signal hydrolase, AiiA. Microbiologyopen 2019, 8, e00797.
- Pang, K.; You, H.; Chen, Y.; Chu, P.; Hu, M.; Shen, J.; Guo, W.; Xie, C.; Lu, B. MagR Alone Is Insufficient to Confer Cellular Calcium Responses to Magnetic Stimulation. Front. Neural Circuits 2017, 11, 11.
- Long, X.; Ye, J.; Zhao, D.; Zhang, S.-J. Magnetogenetics: Remote non-invasive magnetic activation of neuronal activity with a magnetoreceptor. Sci. Bull. 2015, 60, 2107–2119.
- Meister, M. Physical limits to magnetogenetics. eLife 2016, 5, e17210.
- Winklhofer, M.; Mouritsen, H. A room-temperature ferrimagnet made of metallo-proteins? bioRxiv 2016, 8, e094607.
- Xiao, D.-W.; Hu, W.-H.; Cai, Y.; Zhao, N. Magnetic Noise Enabled Biocompass. Phys. Rev. Lett. 2020, 124, 128101.
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers—The database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010, 38, D750–D753.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
2 times
(View History)
Update Date:
23 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




