
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Eleonora Cominelli | + 3485 word(s) | 3485 | 2021-09-29 08:24:08 |
Video Upload Options
Phytic acid has two main roles in plant tissues: storage of phosphorus and regulation of different cellular processes. From a nutritional point of view, it is considered an antinutritional compound because, being a cation chelator, its presence reduces mineral bioavailability from the diet. In recent decades, the development of low phytic acid (lpa) mutants has been an important goal for nutritional seed quality improvement, mainly in cereals and legumes. Different lpa mutations affect phytic acid biosynthetic genes. However, other lpa mutations isolated so far, affect genes coding for three classes of transporters: a specific group of ABCC type vacuolar transporters, putative sulfate transporters, and phosphate transporters.
1. Introduction
2. PA-MRP Transporters
| Class of Transporters | Species | Gene | Phytozome/Genbank/Ensembl Accession Number | Origin of Mutation | Mutation | Reference |
|---|---|---|---|---|---|---|
| MRP | Zea mays | ZmMRP4/ZmABCC4 | EF586878 | EMS | lpa1-1 | [35] |
| lpa1-241 | [36] | |||||
| lpa1-7 | [37] | |||||
| T-DNA insertion | lpa1-mum1 | [24] | ||||
| Embryo specific:RNAi | Ole::MRP4 Glb::MRP4 | [24] | ||||
| Oryza sativa | OsMRP5/OsABCC13 | LOC_Os03g04920 | γ rays + sodium azide | Os-lpa-XS110-2 | [38] | |
| Os-lpa-XS110-3 | [39] | |||||
| T-DNA insertion | 4A-02500 | [39] | ||||
| Embryo specific amiRNA | Ami-MRP5 | [40] | ||||
| Triticum aestivum | TaABCC13-4B | TraesCS4B02G343800 | Constitutive RNAi | TaABCC13 RNAi | [33] | |
| TaABCC13-4D | TraesCS4D02G339000 | |||||
| TaABCC13-5A* | TraesCS5A02G512500 | |||||
| Glycine max | GmMRP3/GmABCC1 | Glyma.03G167800 | EMS | CX1834 | [29][30][31][41] | |
| GmMRP19/GmABCC2 | Glyma.19G169000 | |||||
| GmMRP13/GmABCC3 | Glyma.13G127500 | no reported mutant | no reported mutant | [32] | ||
| Phaseolus vulgaris | PvMRP1/PvABCC1 | Phvul.001G165500 | EMS | lpa1 | [32][42] | |
| lpa12 | [43] | |||||
| PvMRP2/PvABCC2 | Phvul.007G153800 | no reported mutant | no reported mutant | [32] | ||
| SULTR | Oryza sativa | OsSULTR3;3 | LOC_Os04g55800 | γ rays | Oslpa-MH86-1 Os-lpa-Z9B-1 |
[28] [19] |
| OsSULTR3;4 | LOC_Os06g05160 | retrotransposon Tos-17 insertion | spdt-1, spdt-2, spdt-3 | [20] | ||
| Hordeum vulgare | Hvst | HORVU2Hr1G113050 | sodium azide | lpa1-1(M422) | [44] [19] |
|
| Pht | Oryza sativa | OsPht1;4 | LOC_Os04g10750 | retrotransposon Tos-17 insertion | ospt4-1 (NE1260) ospt4-2 (SHIP_ZSF6267) RNAi |
[21] [22] |
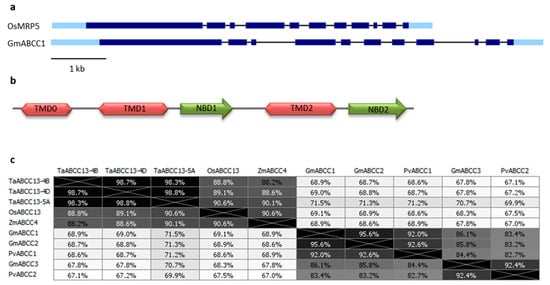
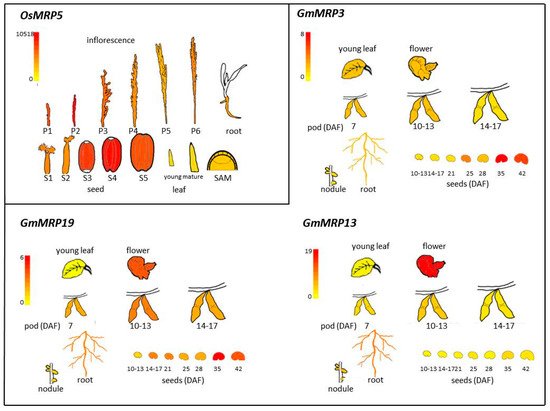
3. SULTR3.3 and SULTR3.4 Transporters Involved in PA Metabolism
| SULTR Group. | Species | Gene Name | Phytozome Accession Number |
|---|---|---|---|
| SULTR3;3 | Zea mays | ZmSULTR3;3 | GRMZM2G395114 |
| Phaseolus vulgaris | PvSULTR3;3 | Phvul.002G095300 | |
| Glycine max | GmSULTR3;3a | Glyma.20G017100 | |
| GmSULTR3;3b | Glyma.07G218900 | ||
| SULTR3;4 | Zea mays | ZmSULTR3;4 | GRMZM2G444801 |
| Phaseolus vulgaris | PvSULTR3;4a | Phvul.005G171800 | |
| PvSULTR3;4b | Phvul.010G151000 | ||
| Glycine max | GmSULTR3;4a | Glyma.07G006500 | |
| GmSULTR3;4b | Glyma.08G207100 | ||
| GmSULTR3;4c | Glyma.13G360000 | ||
| GmSULTR3;4d | Glyma.15G014000 |
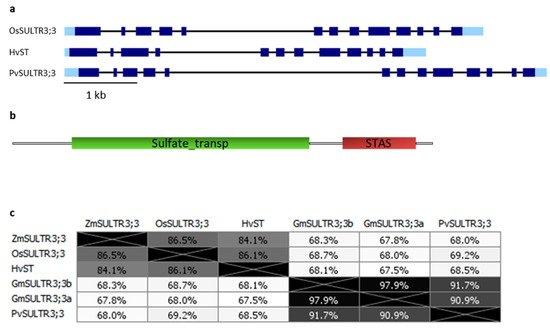
3.1. SULTR3;3
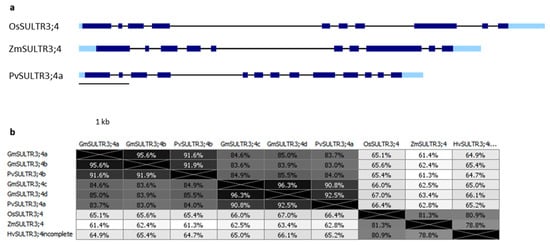
3.2. SULTR3;4
4. OsPht1;4 Phosphate Transporter
The genomic sequence is characterized by the presence of a single exon (Figure 5a) and the protein, 538 aa long, by a major facilitator superfamily domain, characteristic of different transporters, including phosphate transporters (Figure 5b).The OsPT4 gene is mainly expressed in roots, flag leaves and embryos, and its expression is increased in response to prolonged P starvation conditions in shoots and roots, where the signal is specifically localized to the exodermis. The protein is localized to the plasma membrane, as shown in the protoplast system and it is a functional Pi influx transporter, able to complement a yeast mutant defective in Pi uptake and to facilitate the increased accumulation of Pi in Xenopus oocytes.

References
- Sparvoli, F.; Cominelli, E. Seed biofortification and phytic acid reduction: A conflict of interest for the plant? Plants 2015, 4, 728–755.
- Coelho, C.; Tsai, S.; Vitorello, V. Dynamics of inositol phosphate pools (tris-, tetrakis- and pentakisphosphate) in relation to the rate of phytate synthesis during seed development in common bean (Phaseolus vulgaris). J. Plant Physiol. 2005, 162, 1–9.
- Hatzack, F.; Johansen, K.; Rasmussen, S. Nutritionally relevant parameters in low-phytate barley (Hordeum vulgare L.) grain mutants. J. Agric. Food Chem. 2000, 48, 6074–6080.
- Lin, L.; Ockenden, I.; Lott, J. The concentrations and distribution of phytic acid-phosphorus and other mineral nutrients in wild-type and low phytic acid1-1 (lpa1-1) corn (Zea mays L.) grains and grain parts. Can. J. Bot. 2005, 83, 131–141.
- Ockenden, I.; Dorsch, J.; Reid, M.; Lin, L.; Grant, L.; Raboy, V.; Lott, J. Characterization of the storage of phosphorus, inositol phosphate and cations in grain tissues of four barley (Hordeum vulgare L.) low phytic acid genotypes. Plant Sci. 2004, 167, 1131–1142.
- Regvar, M.; Eichert, D.; Kaulich, B.; Gianoncelli, A.; Pongrac, P.; Vogel-Mikus, K.; Kreft, I. New insights into globoids of protein storage vacuoles in wheat aleurone using synchrotron soft X-ray microscopy. J. Exp. Bot. 2011, 62, 3929–3939.
- Krishnan, H. Preparative procedures markedly influence the appearance and structural integrity of protein storage vacuoles in soybean seeds. J. Agric. Food Chem. 2008, 56, 2907–2912.
- O’Dell, B.L.; de Boland, A.R.; Koirtyohann, S.T. Distribution of phytate and nutritionally important elements among the morphological components of cereal grains. J. Agric. Food Chem. 1972, 20, 718–721.
- Ariza-Nieto, M.; Blair, M.; Welch, R.; Glahn, R. Screening of iron bioavailability patterns in eight bean (Phaseolus vulgaris L.) genotypes using the caco-2 cell in vitro model. J. Agric. Food Chem. 2007, 55, 7950–7956.
- Raboy, V. myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 2003, 64, 1033–1043.
- Raboy, V. Seeds for a better future: ‘low phytate’, grains help to overcome malnutrition and reduce pollution. Trends Plant Sci. 2001, 6, 458–462.
- Schlemmer, U.; Frølich, W.; Prieto, R.M.; Grases, F. Phytate in foods and significance for humans: Food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 2009, 53 (Suppl. 2), S330–S375.
- Leytem, A.B.; Maguire, R.O. Environmental implications of inositol phosphates in animal manures. In Inositol Phosphates: Linking Agriculture and the Environment; Turner, B.L., Richardson, A.E., Mullaney, E.J., Eds.; CAB International: Wallingford, CT, USA; Oxfordshire, UK, 2007; pp. 150–168.
- Raboy, V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009, 177, 281–296.
- Martinoia, E. Vacuolar transporters—Companions on a longtime journey. Plant Physiol. 2018, 176, 1384–1407.
- Nagy, R.; Grob, H.; Weder, B.; Green, P.; Klein, M.; Frelet-Barrand, A.; Schjoerring, J.; Brearley, C.; Martinoia, E. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 2009, 284, 33614–33622.
- Colombo, F.; Paolo, D.; Cominelli, E.; Sparvoli, F.; Nielsen, E.; Pilu R. MRP transporters and low phytic acid mutants in major crops: main pleiotropic effects and future perspectives. Front Plant Sci. 2020, 11,1301. doi: 10.3389/fpls.2020.01301. eCollection 2020.
- Takahashi, H.; Buchner, P.; Yoshimoto, N.; Hawkesford, M.J.; Shiu, S.H. Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2011, 2, 119.
- Ye, H.; Zhang, X.; Broughton, S.; Westcott, S.; Wu, D.; Lance, R.; Li, C. A nonsense mutation in a putative sulphate transporter gene results in low phytic acid in barley. Funct. Integr. Genom. 2011, 11, 103–110.
- Yamaji, N.; Takemoto, Y.; Miyaji, T.; Mitani-Ueno, N.; Yoshida, K.T.; Ma, J.F. Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 2017, 541, 92–95.
- Zhang, F.; Sun, Y.; Pei, W.; Jain, A.; Sun, R.; Cao, Y.; Wu, X.; Jiang, T.; Zhang, L.; Fan, X.; et al. Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 2015, 82, 556–569.
- Ye, Y.; Yuan, J.; Chang, X.; Yang, M.; Zhang, L.; Lu, K.; Lian, X. The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS ONE 2015, 10, e0126186.
- Hwang, J.U.; Song, W.Y.; Hong, D.; Ko, D.; Yamaoka, Y.; Jang, S.; Yim, S.; Lee, E.; Khare, D.; Kim, K.; et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol. Plant 2016, 9, 338–355.
- Shi, J.; Wang, H.; Schellin, K.; Li, B.; Faller, M.; Stoop, J.; Meeley, R.; Ertl, D.; Ranch, J.; Glassman, K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 2007, 25, 930–937.
- Gaedeke, N.; Klein, M.; Kolukisaoglu, U.; Forestier, C.; Muller, A.; Ansorge, M.; Becker, D.; Mamnun, Y.; Kuchler, K.; Schulz, B.; et al. The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 2001, 20, 1875–1887.
- Klein, M.; Perfus-Barbeoch, L.; Frelet, A.; Gaedeke, N.; Reinhardt, D.; Mueller-Roeber, B.; Martinoia, E.; Forestier, C. The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. Plant J. 2003, 33, 119–129.
- Suh, S.J.; Wang, Y.F.; Frelet, A.; Leonhardt, N.; Klein, M.; Forestier, C.; Mueller-Roeber, B.; Cho, M.H.; Martinoia, E.; Schroeder, J.I. The ATP binding cassette transporter AtMRP5 modulates anion and calcium channel activities in Arabidopsis guard cells. J. Biol. Chem. 2007, 282, 1916–1924.
- Liu, Q.; Xu, X.; Ren, X.; Fu, H.; Wu, D.; Shu, Q. Generation and characterization of low phytic acid germplasm in rice (Oryza sativa L.). Theor. Appl. Genet. 2007, 114, 803–814.
- Maroof, M.; Glover, N.; Biyashev, R.; Buss, G.; Grabau, E. Genetic basis of the low-phytate trait in the soybean line CX1834. Crop Sci. 2009, 49, 69–76.
- Gillman, J.; Pantalone, V.; Bilyeu, K. The low phytic acid phenotype in soybean line CX1834 is due to mutations in two homologs of the maize low phytic acid gene. Plant Genome 2009, 2, 179–190.
- Gillman, J.; Baxter, I.; Bilyeu, K. Phosphorus partitioning of soybean lines containing different mutant alleles of two soybean seed-specific adenosine triphosphate-binding cassette phytic acid transporter paralogs. Plant Genome 2013, 6.
- Panzeri, D.; Cassani, E.; Doria, E.; Tagliabue, G.; Forti, L.; Campion, B.; Bollini, R.; Brearley, C.A.; Pilu, R.; Nielsen, E.; et al. A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 2011, 191, 70–83.
- Bhati, K.K.; Alok, A.; Kumar, A.; Kaur, J.; Tiwari, S.; Pandey, A.K. Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 2016, 67, 4379–4389.
- Boncompagni, E.; Orozco-Arroyo, G.; Cominelli, E.; Gangashetty, P.I.; Grando, S.; Kwaku Zu, T.T.; Daminati, M.G.; Nielsen, E.; Sparvoli, F. Antinutritional factors in pearl millet grains: Phytate and goitrogens content variability and molecular characterization of genes involved in their pathways. PLoS ONE 2018, 13, e0198394.
- Raboy, V.; Gerbasi, P.F.; Young, K.A.; Stoneberg, S.D.; Pickett, S.G.; Bauman, A.T.; Murthy, P.P.; Sheridan, W.F.; Ertl, D.S. Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 2000, 124, 355–368.
- Pilu, R.; Panzeri, D.; Gavazzi, G.; Rasmussen, S.K.; Consonni, G.; Nielsen, E. Phenotypic, genetic and molecular characterization of a maize low phytic acid mutant (lpa241). Theor. Appl. Genet. 2003, 107, 980–987.
- Cerino Badone, F.; Amelotti, M.; Cassani, E.; Pilu, R. Study of Low Phytic Acid1-7 (lpa1-7), a New ZmMRP4 Mutation in Maize. J. Hered. 2012, 103, 598–605.
- Liu, K.; Peterson, K.; Raboy, V. Comparison of the phosphorus and mineral concentrations in bran and abraded kernel fractions of a normal barley (Hordeum vulgare) cultivar versus four low phytic acid isolines. J. Agric. Food Chem. 2007, 55, 4453–4460.
- Xu, X.; Zhao, H.; Liu, Q.; Frank, T.; Engel, K.; An, G.; Shu, Q. Mutations of the multi-drug resistance-associated protein ABC transporter gene 5 result in reduction of phytic acid in rice seeds. Theor. Appl. Genet. 2009, 119, 75–83.
- Li, W.; Zhao, H.; Pang, W.; Cui, H.; Poirier, Y.; Shu, Q. Seed-specific silencing of OsMRP5 reduces seed phytic acid and weight in rice. Transgenic Res. 2014, 23, 585–599.
- Wilcox, J.; Premachandra, G.; Young, K.; Raboy, V. Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Sci. 2000, 40, 1601–1605.
- Campion, B.; Sparvoli, F.; Doria, E.; Tagliabue, G.; Galasso, I.; Fileppi, M.; Bollini, R.; Nielsen, E. Isolation and characterisation of an lpa (low phytic acid) mutant in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2009, 118, 1211–1221.
- Cominelli, E.; Confalonieri, M.; Carlessi, M.; Cortinovis, G.; Daminati, M.G.; Porch, T.G.; Losa, A.; Sparvoli, F. Phytic acid transport in Phaseolus vulgaris: A new low phytic acid mutant in the PvMRP1 gene and study of the PvMRPs promoters in two different plant systems. Plant Sci. 2018, 270, 1–12.
- Zhao, H.; Liu, Q.; Ren, X.; Wu, D.; Shu, Q. Gene identification and allele-specific marker development for two allelic low phytic acid mutations in rice (Oryza sativa L.). Mol. Breed. 2008, 22, 603–612.
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854.
- Gene Structure Display Server. Available online: http://gsds.cbi.pku.edu.cn/ (accessed on 20 November 2019).
- Patel, R.; Nahal, H.; Breit, R.; Provart, N. BAR expressolog identification: Expression profile similarity ranking of homologous genes in plant species. Plant J. 2012, 71, 1038–1050.
- Bhati, K.; Aggarwal, S.; Sharma, S.; Mantri, S.; Singh, S.; Bhalla, S.; Kaur, J.; Tiwari, S.; Roy, J.; Tuli, R.; et al. Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci. 2014, 224, 74–85.
- Latrasse, D.; Jegu, T.; Meng, P.; Mazubert, C.; Hudik, E.; Delarue, M.; Charon, C.; Crespi, M.; Hirt, H.; Raynaud, C.; et al. Dual function of MIPS1 as a metabolic enzyme and transcriptional regulator. Nucleic Acids Res. 2013, 41, 2907–2917.
- Donahue, J.; Alford, S.; Torabinejad, J.; Kerwin, R.; Nourbakhsh, A.; Ray, W.; Hernick, M.; Huang, X.; Lyons, B.; Hein, P.; et al. The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 2010, 22, 888–903.
- Chen, H.; Xiong, L. myo-inositol-1-phosphate synthase is required for polar auxin transport and organ development. J. Biol. Chem. 2010, 285, 24238–24247.
- Sato, Y.; Yazawa, K.; Yoshida, S.; Tamaoki, M.; Nakajima, N.; Iwai, H.; Ishii, T.; Satoh, S. Expression and functions of myo-inositol monophosphatase family genes in seed development of Arabidopsis. J. Plant Res. 2011, 124, 385–394.
- Nourbakhsh, A.; Collakova, E.; Gillaspy, G.E. Characterization of the inositol monophosphatase gene family in Arabidopsis. Front. Plant Sci. 2014, 5, 725.
- Sweetman, D.; Stavridou, I.; Johnson, S.; Green, P.; Caddick, S.; Brearley, C. Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase 4 (AtITPK4) is an outlier to a family of ATP-grasp fold proteins from Arabidopsis. FEBS Lett. 2007, 581, 4165–4171.
- Xia, H.; Brearley, C.; Elge, S.; Kaplan, B.; Fromm, H.; Mueller-Roeber, B. Arabidopsis inositol polyphosphate 6-/3-kinase is a nuclear protein that complements a yeast mutant lacking a functional ArgR-Mcm1 transcription complex. Plant Cell 2003, 15, 449–463.
- Zhang, Z.B.; Yang, G.; Arana, F.; Chen, Z.; Li, Y.; Xia, H.J. Arabidopsis inositol polyphosphate 6-/3-kinase (AtIpk2beta) is involved in axillary shoot branching via auxin signaling. Plant Physiol. 2007, 144, 942–951.
- Munnik, T.; Vermeer, J. Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ. 2010, 33, 655–669.
- Zhao, H.; Frank, T.; Tan, Y.; Zhou, C.; Jabnoune, M.; Arpat, A.B.; Cui, H.; Huang, J.; He, Z.; Poirier, Y.; et al. Disruption of OsSULTR3;3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytol. 2016, 211, 926–939.
- Shoemaker, R.C.; Polzin, K.; Labate, J.; Specht, J.; Brummer, E.C.; Olson, T.; Young, N.; Concibido, V.; Wilcox, J.; Tamulonis, J.P.; et al. Genome duplication in soybean (Glycine subgenus soja). Genetics 1996, 144, 329–338.
- Yamaji, N.; Ma, J.F. Node-controlled allocation of mineral elements in Poaceae. Curr. Opin. Plant Biol. 2017, 39, 18–24.
- Liu, F.; Chang, X.; Ye, Y.; Xie, W.; Wu, P.; Lian, X. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 2011, 4, 1105–1122.




