| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | xiaoxu shen | + 2395 word(s) | 2395 | 2021-08-19 04:34:53 | | | |
| 2 | Rita Xu | -1 word(s) | 2394 | 2021-09-02 09:52:06 | | | | |
| 3 | Rita Xu | Meta information modification | 2394 | 2021-09-02 12:01:27 | | |
Video Upload Options
A novel circular PPP1R13B RNA promotes chicken skeletal muscle satellite cell proliferation and differentiation via targeting miR-9-5p and activating IGF/PI3K/AKT signaling pathway.
1. Introduction
2. Circular PPP1R13B RNA is Differentially Expressed between FMGB and SMGL
From our previous sequencing data (in the Sequence Read Archive database with accession number PRJNA516545), we found that circPPP1R13B was one of the differentially expressed circRNAs among FMGB and SMGL at all four time points (E10, E13, E16 and E19). Furthermore, we identified that circPPP1R13B came from the exon 2-4 of PPP1R13B gene, and the junction sequence which connects the head of exon 2 and the tail of exon 4. Our results also showed that PPP1R13B mRNA was digested by RNase R instead of circPPP1R13B, and in the reverse transcription reaction, random primers promoted circPPP1R13B cDNA synthesis more than oligo d (T). These results confirmed the circular form of circPPP1R13B. Thereafter, the expression of circPPP1R13B in the chicken was identified by qRT-PCR. The results were similar to the sequencing data, which showed that the expression of circPPP1R13B was higher in the FMGB as compared to that in the SMGL at E10, E13 and E16. Furthermore, we found that circPPP1R13B was also expressed in the fat, liver, spleen, heart, lung, brain, and intestine tissues, however, its expression was higher in skeletal muscle, heart and fat tissues than the other tissues .
In order to explore the potential function of circPPP1R13B in chicken skeletal muscle development, siRNAs and overexpression vector were used to control the expression of circPPP1R13B. The results showed that siRNAs, especially siRNA1 could significantly reduce the expression of circPPP1R13B, therefore siRNA1 was chosen for further experiments (named si-circPPP1R13B). Besides, overexpression vector significantly increased the expression of circPPP1R13B.
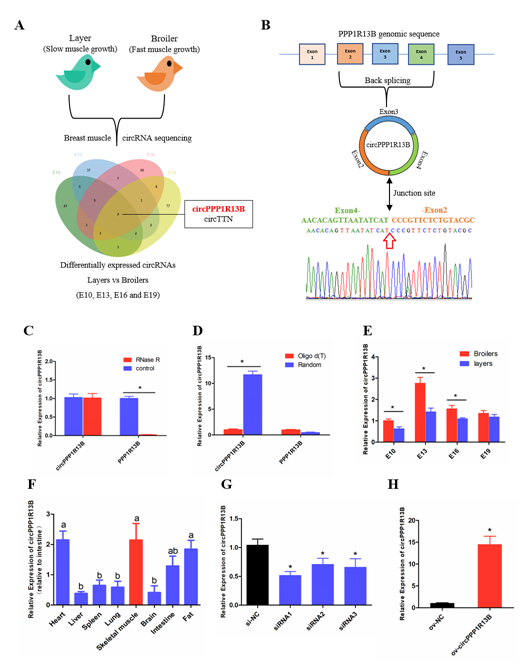
Figure 1. The selection, identification and expression pattern of circPPP1R13B. (A) differentially expressed circRNA analysis revealed that circPPP1R13B was chosen as a candidate circRNA, (B) The formation diagram and junction sequence of circPPP1R13B. (C) The expression of circPPP1R13B and linear PPP1R13B under RNase R treatment. (D) The expression of circPPP1R13B and linear PPP1R13B in reverse transcription samples with different primers. (E) The expression of circPPP1R13B in breast muscles of broilers and layers at four embryonic stage. (F) The expression of circPPP1R13B in different tissues of 4-day-old chickens. (G) Knock down efficiency of circPPP1R13B in SMSCs by exogenous siRNAs. (H) Overexpression efficiency of circPPP1R13B in SMSCs by overexpress vectors. Asterisk and different letters indicate significant differences from the respective control (* p < 0.05).
3. CircPPP1R13B Interacts with miR-9-5p in Chicken SMSCs
Previous studies indicated that a circRNA functions as a miRNA sponge. To verify whether circPPP1R13B sponges the miRNAs, RNAhybrid software was used for miRNA target prediction. The results showed that eighteen miRNAs directly target circPPP1R13B, among them, miR-9-5p was our focus due to the finding of our previous study which reported that, miR-9-5p could regulate proliferation and differentiation in chicken SMSCs. To observe the relation between circPPP1R13B and miR-9-5p, dual luciferase reporter vector which contains wild type or mutant type binding site of miR-9-5p were constructed, and co-transfected with miR-9-5p or negative mimic into the DF-1 cells. The results showed that miR-9-5p significantly reduced the luciferase of 3′ UTR which contain miR-9-5p wild type binding site, but had no effect on the luciferase with mutant type binding site. In addition, knockdown of circPPP1R13B significantly elevated miR-9-5p RNA level, whereas overexpression of circPPP1R13B significantly decreased the expression of miR-9-5p. These results suggested that circPPP1R13B acts as a sponge of miR-9-5p.
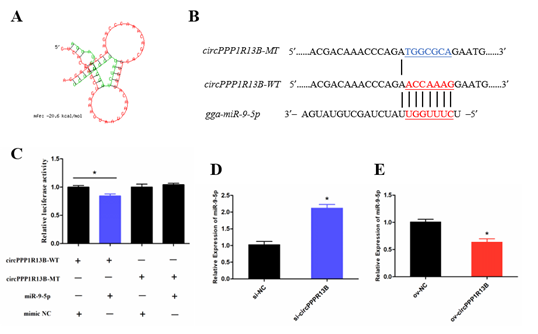
Figure 2. CircPPP1R13B directly targets miR-9-5p. (A) the target site of miR-9-5p on circPPP1R13B predicted by RNAhybrid software. (B) wild type and mutant type of miR-9-5p targeting site used for dual luciferase reporter vector construction. (C) Relative luciferase activity of wild type or mutant type reporter vectors co-transfected with miR-9-5p mimic or negative mimic. (D) The expression of miR-9-5p after circPPP1R13B knockdown in chicken SMSCs. (E) The expression of miR-9-5p after circPPP1R13B overexpression in chicken SMSCs. Asterisk indicate significant differences from the respective control (* p < 0.05).
4. CircPPP1R13B Regulates IGF2BP3 via Targeting miR-9-5p in Chicken SMSCs
IGF2BP3 is a direct target gene of miR-9-5p, miR-9-5p inhibits the expression levels of IGF2BP3 and blocks the activity of downstream IGF/PI3K/AKT signaling pathway. To verify whether circPPP1R13B regulates the expression of IGF2BP3 and downstream signaling pathway via targeting miR-9-5p, we performed co-transfection experiment, and the results showed that, the knockdown of circPPP1R13B decreased the RNA level of IGF2BP3, whereas the overexpression of miR-9-5p inhibited the mRNA expression of IGF2BP3 and co-overexpression of both miR-9-5p and circPPP1R13B restored the RNA level of IGF2BP3. The protein level of IGF2BP3 also showed similar trend as the RNA level results. Furthermore, the key regulator of IGF/PI3K/AKT signaling pathway (phosphorylation of AKT1), the results showed the same trend after co-transfection.
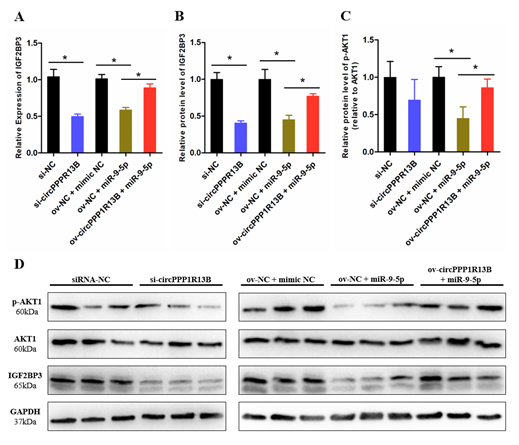
Figure 3. CircPPP1R13B regulates IGF2BP3 mediated IGF/PI3K/AKT signaling via targeting miR-9-5p. (A) Relative mRNA expression of IGF2BP3 following circPPP1R13B knockdown or co-overexpression with miR-9-5p. (B) Relative protein level of IGF2BP3 following circPPP1R13B knockdown or co-overexpression with miR-9-5p. (C) Relative protein phosphorylation level of AKT1 following circPPP1R13B knockdown or co-overexpression with miR-9-5p. (D) Protein levels of IGF2BP3, p-AKT1, AKT1, and GAPDH were determined by western blot analysis. Asterisk indicate significant differences from the respective control (* p < 0.05).
5. CircPPP1R13B Eliminates the Inhibition Effect of miR-9-5p on Chicken SMSC Proliferation
To explore the role of circPPP1R13B in chicken SMSC proliferation, cell cycle analysis and EdU assay were performed in the circPPP1R13B knockdown SMSCs. Cell cycle analysis results showed that the knockdown of circPPP1R13B makes the SMSCs stagnant during the G0/G1 phase, and reduce the number of cells in division (G2/M) phase. EdU assay results showed that, the knockdown of circPPP1R13B decreased the number of EdU labeled proliferating cells. These results indicated that circPPP1R13B may play positive roles in SMSC proliferation.
Furthermore, we explored the relation between circPPP1R13B and miR-9-5p in SMSC proliferation. Cell cycle analysis results showed that, the overexpression of miR-9-5p reduced the population of G2/M phase cells and increased the population of G0/G1 phase cells, whereas the effect of miR-9-5p on SMSC cell cycle was alleviated by co-overexpressed miR-9-5p and circPPP1R13B. Similar to cell cycle analysis results, the EdU assay showed that the overexpression of miR-9-5p decreased the number of EdU positive cells, whereas the inhibition role of miR-9-5p on cell proliferation was eliminated by the co-overexpression with circPPP1R13B. Thus, these results suggested that circPPP1R13B eliminated the inhibition effect of miR-9-5p on chicken SMSC proliferation.
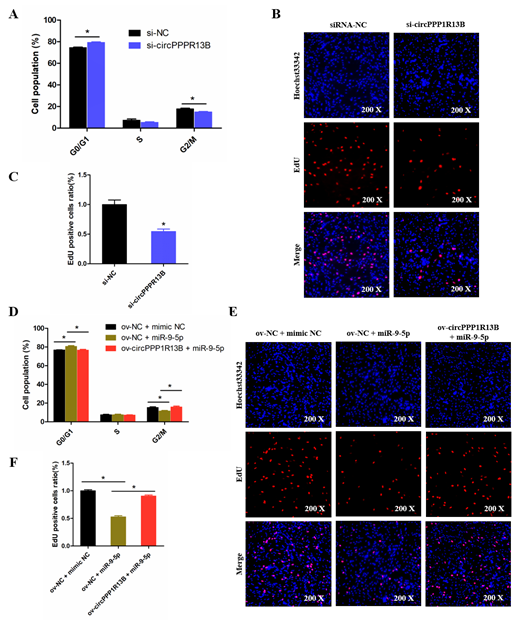
Figure 4. CircPPP1R13B regulates chicken SMSCs proliferation by targeting miR-9-5p. (A) Cell cycle analysis of SMSCs following circPPP1R13B knockdown. (B) EdU analysis plot of SMSCs following circPPP1R13B knockdown. (C) Relative EdU positive cells following circPPP1R13B knockdown. (D) Cell cycle analysis of SMSCs following co-overexpress circPPP1R13B with miR-9-5p. (E) EdU analysis plot of SMSCs following co-overexpress circPPP1R13B with miR-9-5p. (F) Relative EdU positive cells following co-overexpress circPPP1R13B with miR-9-5p. Asterisk indicate significant differences from the respective control (* p < 0.05).
6. CircPPP1R13B Attenuates the Inhibition Effect of miR-9-5p on Chicken SMSC Differentiation
To observe the function of circPPP1R13B on chicken SMSC differentiation, qRT-PCR, Western blot, and Immunofluorescence were performed in the circPPP1R13B knockdown SMSCs. qRT-PCR results showed that, the knockdown of circPPP1R13B significantly inhibited the RNA level of skeletal muscle development marker genes MyoG, Myogenic Differentiation 1 (MyoD1) and MyHC, Western blot result was similar to qRT-PCR data, which showed the protein level of MyoG were decreased by circPPP1R13B knockdown. In addition, the knockdown of circPPP1R13B inhibits the level of myotube component protein MyHC, which marks the formation of myotubes. These results showed that circPPP1R13B may play a positive role in chicken SMSC differentiation.
Furthermore, we explored the relation between circPPP1R13B and miR-9-5p on SMSC differentiation, and we observed that, the overexpression of miR-9-5p decreased the RNA level and protein level of skeletal muscle development marker genes, whereas the inhibitory effect of miR-9-5p on SMSC differentiation was prevented after the co-transfection with circPPP1R13B. Furthermore, miR-9-5p inhibited myotube formation, co-overexpression of the circPPP1R13B and miR-9-5p prevented the inhibition of miR-9-5p on myotube formation. Altogether, these results suggested circPPP1R13B attenuated the inhibition effect of miR-9-5p on chicken SMSC differentiation.
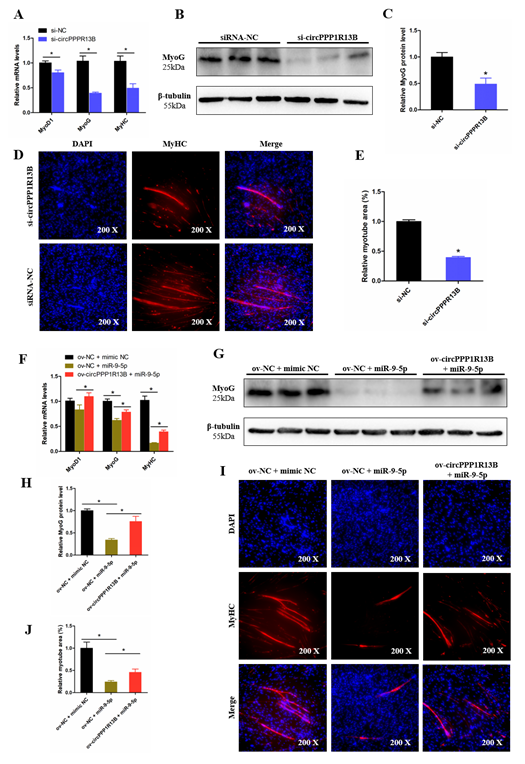
Figure 5. CircPPP1R13B regulates chicken SMSCs differentiation by targeting miR-9-5p. (A) Relative expression of muscle cell differentiation marker genes following circPPP1R13B knockdown. (B) Protein level of MyoG and β-tubulin observed by western blot analysis. (C) Relative protein level of MyoG following circPPP1R13B knockdown. (D) Images of MyHC staining of SMSCs following circPPP1R13B knockdown. (E) Relative myotube area of SMSCs following circPPP1R13B knockdown. (F) Relative expression of muscle cell differentiation marker genes following circPPP1R13B co-overexpress circPPP1R13B with miR-9-5p. (G) Protein level of MyoG and β-tubulin observed by western blot analysis. (H) Relative protein level of MyoG following circPPP1R13B co-overexpress circPPP1R13B with miR-9-5p. (I) Images of MyHC staining of SMSCs following circPPP1R13B co-overexpress circPPP1R13B with miR-9-5p. (J) Relative myotube area of SMSCs following circPPP1R13B co-overexpress circPPP1R13B with miR-9-5p. Asterisk indicate significant differences from the respective control (* p < 0.05).
7. Conclusion
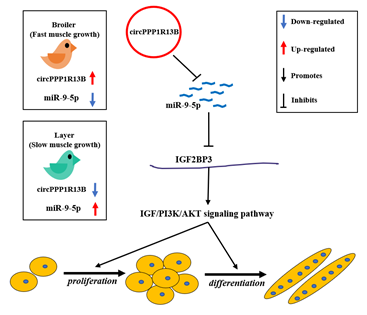
In conclusion, we found that a novel circRNA circPPP1R13B was highly expressed in the breast muscle of FMGB compared to SMGL, and confirmed that circPPP1R13B could promote SMSC proliferation and differentiation in chicken. Furthermore, we established that circPPP1R13B targets SMGL enriched miR-9-5p to repress the negative effects of miR-9-5p on chicken SMSC proliferation and differentiation. Therefore, the results obtained in this study revealed that circPPP1R13B could act as a potential target factor in chicken molecular breeding to improve skeletal muscle growth in broilers.
Figure 6. A schematic model of circPPP1R13B regulates chicken SMSC proliferation and differentiation. CircPPP1R13B highly expressed in the breast muscle of FMGB compared to that in SMGL, whereas miR-9-5p highly expressed in the breast muscle of SMGL compared to FMGB. Furthermore, circPPP1R13B promotes chicken SMSC proliferation and differentiation via targets miR-9-5p to active IGF2BP3 mediated IGF/PI3K/AKT signaling pathway.
References
- Yanli Du; Linlin Liu; Yang He; Tengfei Dou; Junjing Jia; Changrong Ge; Endocrine and genetic factors affecting egg laying performance in chickens: a review. British Poultry Science 2020, 61, 538-549, 10.1080/00071668.2020.1758299.
- Domenico Aiello; K. Patel; E. Lasagna; Themyostatingene: an overview of mechanisms of action and its relevance to livestock animals. Animal Genetics 2018, 49, 505-519, 10.1111/age.12696.
- Weijian Li; Rongyang Li; Yinghui Wei; Xueqing Meng; Binbin Wang; Zengkai Zhang; Wangjun Wu; Honglin Liu; Effect of MSTN Mutation on Growth and Carcass Performance in Duroc × Meishan Hybrid Population. Animals 2020, 10, 932, 10.3390/ani10060932.
- Shudai Lin; Congjun Li; Charles Li; Xiquan Zhang; Growth Hormone Receptor Mutations Related to Individual Dwarfism. International Journal of Molecular Sciences 2018, 19, 1433, 10.3390/ijms19051433.
- H.X. Cui; Q.C. Shen; M.Q. Zheng; Y.C. Su; R.C. Cai; Y Yu; X.R. Yang; Z.W. Chen; J Wen; G.P. Zhao; et al. A selection method of chickens with blue-eggshell and dwarf traits by molecular marker-assisted selection. Poultry Science 2019, 98, 3114-3118, 10.3382/ps/pez069.
- Mohammadreza Mohammadabadi; Farhad Bordbar; Just Jensen; Min Du; Wei Guo; Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals. Animals 2021, 11, 835, 10.3390/ani11030835.
- Fabien Le Grand; Michael A Rudnicki; Skeletal muscle satellite cells and adult myogenesis. Current Opinion in Cell Biology 2007, 19, 628-633, 10.1016/j.ceb.2007.09.012.
- Michael Taylor; Simon M. Hughes; Mef2 and the skeletal muscle differentiation program. Seminars in Cell & Developmental Biology 2017, 72, 33-44, 10.1016/j.semcdb.2017.11.020.
- Bidossessi Nadège Zanou; Philippe Gailly; Skeletal muscle hypertrophy and regeneration: interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cellular and Molecular Life Sciences 2013, 70, 4117-4130, 10.1007/s00018-013-1330-4.
- Gerald Soslau; Circular RNA (circRNA) was an important bridge in the switch from the RNA world to the DNA world. Journal of Theoretical Biology 2018, 447, 32-40, 10.1016/j.jtbi.2018.03.021.
- H. L. Sanger; G. Klotz; D. Riesner; H. J. Gross; A. K. Kleinschmidt; Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures.. Proceedings of the National Academy of Sciences 1976, 73, 3852-3856, 10.1073/pnas.73.11.3852.
- Wanying Wu; Peifeng Ji; Fangqing Zhao; CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biology 2020, 21, 1-14, 10.1186/s13059-020-02018-y.
- Lasse S. Kristensen; Maria S. Andersen; Lotte V. W. Stagsted; Karoline K. Ebbesen; Thomas B. Hansen; Jørgen Kjems; The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics 2019, 20, 675-691, 10.1038/s41576-019-0158-7.
- Li Li; Yuan Chen; Lu Nie; Xue Ding; Xiao Zhang; Wei Zhao; Xiaoli Xu; Bismark Kyei; Dinghui Dai; Siyuan Zhan; et al.Jiazhong GuoTao ZhongLinjie WangHongping Zhang MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochimica et Biophysica Acta 2019, 1862, 807-821, 10.1016/j.bbagrm.2019.07.001.
- Mengjin Gao; Xue Li; Zuojun Yang; Shuo Zhao; Xingxing Ling; Jingjing Li; Kai Xing; Xiaolong Qi; Xiangguo Wang; Longfei Xiao; et al.Hemin NiYong GuoXihui Sheng circHIPK3 regulates proliferation and differentiation of myoblast through the miR‐7/ TCF12 pathway. Journal of Cellular Physiology 2021, 236, 6793-6805, 10.1002/jcp.30363.
- Jia Liu; Menglu Li; Linghao Kong; Mengwen Cao; Molan Zhang; Yanhong Wang; Chengchuang Song; Xingtang Fang; Hong Chen; Chunlei Zhang; et al. CircARID1A regulates mouse skeletal muscle regeneration by functioning as a sponge of miR‐6368. The FASEB Journal 2021, 35, e21324, 10.1096/fj.202001992r.
- Xuemei Shen; Jia Tang; Rui Jiang; Xiaogang Wang; Zhaoxin Yang; Yongzhen Huang; Xianyong Lan; Chuzhao Lei; Hong Chen; CircRILPL1 promotes muscle proliferation and differentiation via binding miR-145 to activate IGF1R/PI3K/AKT pathway. Cell Death & Disease 2021, 12, 1-14, 10.1038/s41419-021-03419-y.
- H.D. Griffin; C. Goddard; Rapidly growing broiler (meat-type) chickens. Their origin and use for comparative studies of the regulation of growth. International Journal of Biochemistry 1994, 26, 19-28, 10.1016/0020-711x(94)90190-2.
- Xiaoxu Shen; Zihao Liu; Xinao Cao; Haorong He; Shunshun Han; Yuqi Chen; Can Cui; Jing Zhao; Diyan Li; Yan Wang; et al.Qing ZhuHuadong Yin Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. International Journal of Biological Sciences 2019, 15, 2265-2281, 10.7150/ijbs.36412.