
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bishnu P. Regmi | + 2840 word(s) | 2840 | 2021-08-31 05:31:31 | | | |
| 2 | Peter Tang | Meta information modification | 2840 | 2021-09-02 02:58:46 | | |
Video Upload Options
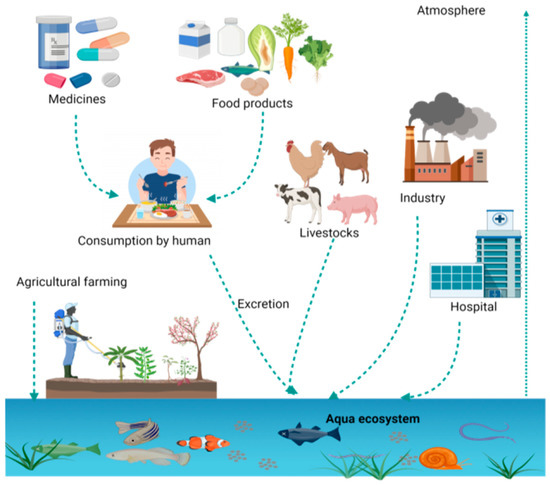
Overuse of antibiotics leads to their circulation in the food chain due to unmanaged discharge. These circulating antibiotics and their residues are a major cause of antimicrobial resistance (AMR), so comprehensive and multifaceted measures aligning with the One Health approach are crucial to curb the emergence and dissemination of antibiotic resistance through the food chain. Different chromatographic techniques and capillary electrophoresis (CE) are being widely used for the separation and detection of antibiotics and their residues from food samples.
1. Introduction
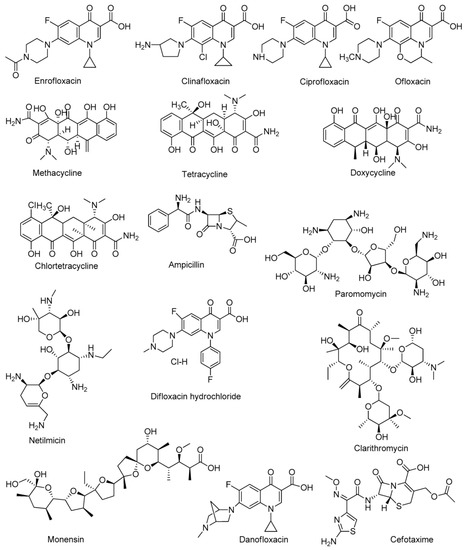
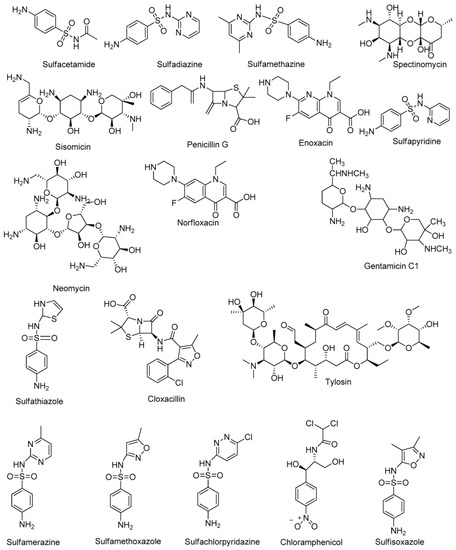
|
Class |
Examples |
References |
|---|---|---|
|
Glycopeptides |
Vancomycin, Teicoplanin, Telavancin, Oritavancin, Dalbavancin |
[12] |
|
Sulfonamides |
Sulfacetamide, Sulfadiazine, Sulfathiazole, Sulfapyridine, Sulfamerazine, Sulfamethazine, Sulfamethoxazole, Sulfasoxazole, Sulfachloropyridazine |
[13] |
|
Tetracyclines |
Tetracycline, Oxytetracycline, Doxycycline, Chlorotetracycline, Methacycline |
[13] |
|
Aminoglycosides |
Amikacin, Paramomycin, Dihydrostreptomycin, Hygromycin, Kanamycin, Netilmycin, Spectinomycin, Sisomycin, Streptomycin, Tobramycin, Gentamicin, Neomycin |
[13] |
|
Β—Lactams |
Amoxicillin, Ampicillin, Cloxacillin, Penicillin G |
[13] |
|
Macrolides |
Erythromycin, Clarithromycin, Tylosin |
[13] |
|
Fluoroquinolones |
Lomefloxacin, Ciprofloxacin, Enroflaxacin, Danofloxacin, Difloxacin hydrochloride, Clinafloxacin |
|
|
Polyethers |
Lasalocid, Salinomycin, Monensin, Narasin, Nigericin |
[14] |
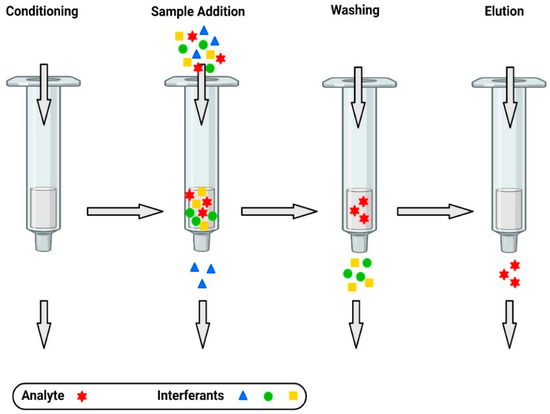
2. Extraction of Antibiotics from Food Samples
3. Separation and Detection of Antibiotics
3.1. Chromatography
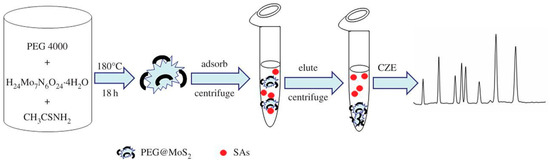
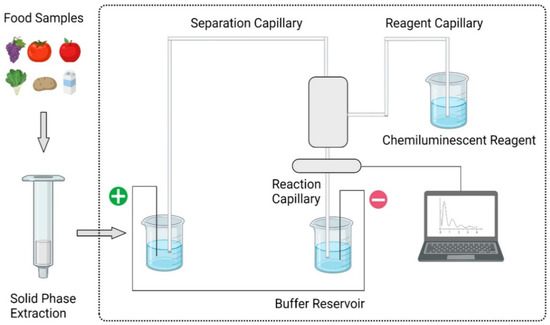
3.2. Capillary Electrophoresis
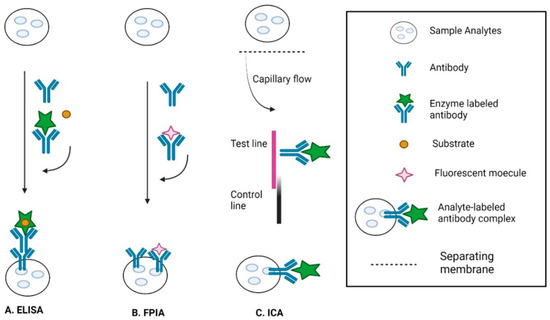
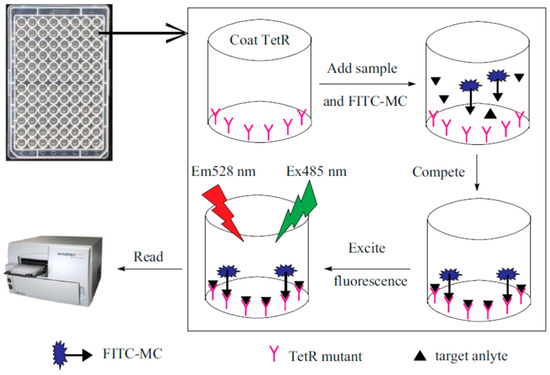
3.3. Immunological Methods
3.4. Surface-Enhanced Raman Spectroscopy (SERS)-Based Methods
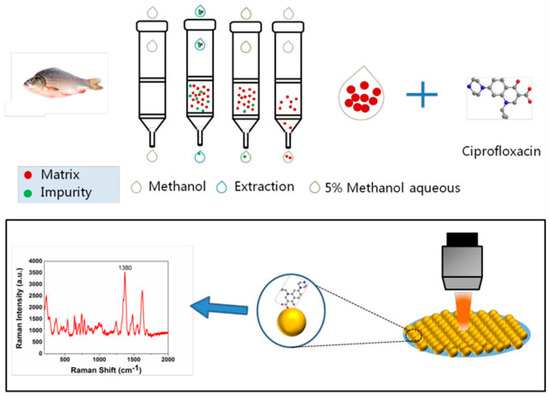
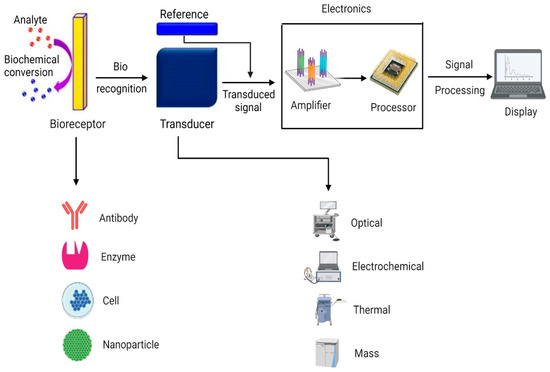
3.5. Biosensors
References
- Chandler, C.I.R. Current Accounts of Antimicrobial Resistance: Stabilisation, Individualisation and Antibiotics as Infrastructure. Palgrave Commun. 2019, 5, 53.
- Angelakis, E.; Merhej, V.; Raoult, D. Related Actions of Probiotics and Antibiotics on Gut Microbiota and Weight Modification. Lancet Infect. Dis. 2013, 13, 889–899.
- Menkem, Z.E.; Ngangom, B.L.; Tamunjoh, S.S.A.; Boyom, F.F. Antibiotic Residues in Food Animals: Public Health Concern. Acta Ecol. Sin. 2019, 39, 411–415.
- Gothwal, R.; Shashidhar, T. Antibiotic Pollution in the Environment: A Review: Antibiotic Pollution in the Environment. Clean Soil Air Water 2015, 43, 479–489.
- Ji, K.; Kho, Y.; Park, C.; Paek, D.; Ryu, P.; Paek, D.; Kim, M.; Kim, P.; Choi, K. Influence of Water and Food Consumption on Inadvertent Antibiotics Intake among General Population. Environ. Res. 2010, 110, 641–649.
- Santos, L.; Ramos, F. Analytical Strategies for the Detection and Quantification of Antibiotic Residues in Aquaculturefishes: A Review. Trends Food Sci. Technol. 2016, 52.
- Joshi, A.; Kim, K.-H. Recent Advances in Nanomaterial-Based Electrochemical Detection of Antibiotics: Challenges and Future Perspectives. Biosens. Bioelectron. 2020, 153, 112046.
- Bueno, I.; Williams-Nguyen, J.; Hwang, H.; Sargeant, J.M.; Nault, A.J.; Singer, R.S. Systematic Review: Impact of Point Sources on Antibiotic-Resistant Bacteria in the Natural Environment. Zoonoses Public Health 2018, 65, e162–e184.
- Rozman, U.; Duh, D.; Cimerman, M.; Turk, S.Š. Hospital Wastewater Effluent: Hot Spot for Antibiotic Resistant Bacteria. J. Water Sanit. Hyg. Dev. 2020, 10, 171–178.
- Petrovich, M.L.; Zilberman, A.; Kaplan, A.; Eliraz, G.R.; Wang, Y.; Langenfeld, K.; Duhaime, M.; Wigginton, K.; Poretsky, R.; Avisar, D.; et al. Microbial and Viral Communities and Their Antibiotic Resistance Genes Throughout a Hospital Wastewater Treatment System. Front. Microbiol. 2020, 11, 153.
- Kumar, A.; Pal, D. Antibiotic Resistance and Wastewater: Correlation, Impact and Critical Human Health Challenges. J. Environ. Chem. Eng. 2018, 6, 52–58.
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80.
- Coates, A.R.; Halls, G.; Hu, Y. Novel Classes of Antibiotics or More of the Same?: New Antibiotic Classes Are Urgently Needed. Br. J. Pharmacol. 2011, 163, 184–194.
- Ha, J.; Song, G.; Ai, L.-F.; Li, J.-C. Determination of Six Polyether Antibiotic Residues in Foods of Animal Origin by Solid Phase Extraction Combined with Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. B 2016, 1017–1018, 187–194.
- Chen, J.; Ying, G.-G.; Deng, W.-J. Antibiotic Residues in Food: Extraction, Analysis, and Human Health Concerns. J. Agric. Food Chem. 2019, 67, 7569–7586.
- Phillips, I. Does the Use of Antibiotics in Food Animals Pose a Risk to Human Health? A Critical Review of Published Data. J. Antimicrob. Chemother. 2003, 53, 28–52.
- Kneebone, J.; Tsang, P.C.W.; Towson, D.H. Rapid Antibiotic Screening Tests Detect Antibiotic Residues in Powdered Milk Products. J. Dairy Sci. 2010, 93, 3961–3964.
- Jammoul, A.; El Darra, N. Evaluation of Antibiotics Residues in Chicken Meat Samples in Lebanon. Antibiotics 2019, 8, 69.
- Farouk, F.; Azzazy, H.M.E.; Niessen, W.M.A. Challenges in the Determination of Aminoglycoside Antibiotics, a Review. Anal. Chim. Acta 2015, 890, 21–43.
- Larsson, D.G.J. Antibiotics in the Environment. Upsala J. Med. Sci. 2014, 119, 108–112.
- Majdinasab, M.; Mishra, R.K.; Tang, X.; Marty, J.L. Detection of Antibiotics in Food: New Achievements in the Development of Biosensors. TrAC Trends Anal. Chem. 2020, 127, 115883.
- Khan, M.Z.H. Recent Biosensors for Detection of Antibiotics in Animal Derived Food. Crit. Rev. Anal. Chem. 2020, 1–11.
- Colombo, R.; Papetti, A. Advances in the Analysis of Veterinary Drug Residues in Food Matrices by Capillary Electrophoresis Techniques. Molecules 2019, 24, 4617.
- Li, W.; Dai, X.; Pu, E.; Bian, H.; Chen, Z.; Zhang, X.; Guo, Z.; Li, P.; Li, H.; Yong, Y.; et al. HLB-MCX-Based Solid-Phase Extraction Combined with Liquid Chromatography–Tandem Mass Spectrometry for the Simultaneous Determination of Four Agricultural Antibiotics (Kasugamycin, Validamycin A, Ningnanmycin, and Polyoxin B) Residues in Plant-Origin Foods. J. Agric. Food Chem. 2020, 68, 14025–14037.
- Di, S.; Yu, J.; Chen, P.; Zhu, G.; Zhu, S. Net-like Mesoporous Carbon Nanocomposites for Magnetic Solid-Phase Extraction of Sulfonamides Prior to Their Quantitation by UPLC-HRMS. Microchim. Acta 2020, 187, 112.
- Alampanos, V.; Samanidou, V.; Papadoyannis, I. Trends in Sample Preparation for the HPLC Determination of Penicillins in Biofluids. J. Appl. Bioanal. 2019, 5, 9–17.
- Sun, Y.; Zhao, J.; Liang, L. Recent Development of Antibiotic Detection in Food and Environment: The Combination of Sensors and Nanomaterials. Microchim. Acta 2021, 188, 21.
- Xu, G.; Zhang, B.; Wang, X.; Li, N.; Zhao, Y.; Liu, L.; Lin, J.-M.; Zhao, R.-S. Porous Covalent Organonitridic Frameworks for Solid-Phase Extraction of Sulfonamide Antibiotics. Mikrochim. Acta 2018, 186, 26.
- Pérez-Rodríguez, M.; Pellerano, R.G.; Pezza, L.; Pezza, H.R. An Overview of the Main Foodstuff Sample Preparation Technologies for Tetracycline Residue Determination. Talanta 2018, 182, 1–21.
- Li, T.; Wang, C.; Xu, Z.; Chakraborty, A. A Coupled Method of On-Line Solid Phase Extraction with the UHPLC‒MS/MS for Detection of Sulfonamides Antibiotics Residues in Aquaculture. Chemosphere 2020, 254, 126765.
- Xu, G.; Dong, X.; Hou, L.; Wang, X.; Liu, L.; Ma, H.; Zhao, R.-S. Room-Temperature Synthesis of Flower-Shaped Covalent Organic Frameworks for Solid-Phase Extraction of Quinolone Antibiotics. Anal. Chim. Acta 2020, 1126, 82–90.
- Betina, V. Bioautography in Paper and Thin-Layer Chromatography and Its Scope in the Antibiotic Field. J. Chromatogr. A 1973, 78, 41–51.
- Zhao, Y.; Wu, R.; Yu, H.; Li, J.; Liu, L.; Wang, S.; Chen, X.; Chan, T.-W.D. Magnetic Solid-Phase Extraction of Sulfonamide Antibiotics in Water and Animal-Derived Food Samples Using Core-Shell Magnetite and Molybdenum Disulfide Nanocomposite Adsorbent. J. Chromatogr. A 2020, 1610, 460543.
- Song, X.; Zhou, T.; Li, J.; Su, Y.; Xie, J.; He, L. Determination of Macrolide Antibiotics Residues in Pork Using Molecularly Imprinted Dispersive Solid-Phase Extraction Coupled with LC-MS/MS. J. Sep. Sci. 2018, 41, 1138–1148.
- Armentano, A.; Summa, S.; Lo Magro, S.; Palermo, C.; Nardiello, D.; Centonze, D.; Muscarella, M. Rapid Method for the Quantification of 13 Sulphonamides in Milk by Conventional High-Performance Liquid Chromatography with Diode Array Ultraviolet Detection Using a Column Packed with Core-Shell Particles. J. Chromatogr. A 2018, 1531, 46–52.
- Hui, W.; Li, Q.; Ma, H.; Wu, M.; Feng, K.; Zhu, H.; Yang, P.; Li, J.; Chen, C.; Yan, K. Rapid Screening for 15 Sulfonamide Residues in Foods of Animal Origin by High-Performance Liquid Chromatography–UV Method. J. Chromatogr. Sci. 2018, 56, 636–643.
- Łukaszewicz, P.; Białk-Bielińska, A.; Dołżonek, J.; Kumirska, J.; Caban, M.; Stepnowski, P. A New Approach for the Extraction of Tetracyclines from Soil Matrices: Application of the Microwave-Extraction Technique. Anal. Bioanal. Chem. 2018, 410, 1697–1707.
- Wang, Z.; Song, X.; Zhou, T.; Bian, K.; Zhang, F.; He, L.; Liu, Q. Simultaneous Determination of Ten Macrolides Drugs in Feeds by High Performance Liquid Chromatography with Evaporation Light Scattering Detection. RSC Adv. 2015, 5, 1491–1499.
- Song, X.; Xie, J.; Zhang, M.; Zhang, Y.; Li, J.; Huang, Q.; He, L. Simultaneous Determination of Eight Cyclopolypeptide Antibiotics in Feed by High Performance Liquid Chromatography Coupled with Evaporation Light Scattering Detection. J. Chromatogr. B 2018, 1076, 103–109.
- Lu, Z.; Deng, F.; He, R.; Tan, L.; Luo, X.; Pan, X.; Yang, Z. A Pass-through Solid-Phase Extraction Clean-up Method for the Determination of 11 Quinolone Antibiotics in Chicken Meat and Egg Samples Using Ultra-Performance Liquid Chromatography Tandem Mass Spectrometry. Microchem. J. 2019, 151, 104213.
- Yu, H.; Jia, Y.; Wu, R.; Chen, X.; Chan, T.-W.D. Determination of Fluoroquinolones in Food Samples by Magnetic Solid-Phase Extraction Based on a Magnetic Molecular Sieve Nanocomposite Prior to High-Performance Liquid Chromatography and Tandem Mass Spectrometry. Anal. Bioanal. Chem. 2019, 411, 2817–2826.
- He, T.; Xu, Z.; Ren, J. Pressure-Assisted Electrokinetic Injection Stacking for Seven Typical Antibiotics in Waters to Achieve Μg/L Level Analysis by Capillary Electrophoresis with UV Detection. Microchem. J. 2019, 146, 1295–1300.
- Moreno-González, D.; Krulišová, M.; Gámiz-Gracia, L.; García-Campaña, A.M. Determination of Tetracyclines in Human Urine Samples by Capillary Electrophoresis in Combination with Field Amplified Sample Injection. Electrophoresis 2018, 39, 608–615.
- Ferreira, T.A.; Flores-Aguilar, J.F.; Santos, E.M.; Rodriguez, J.A.; Ibarra, I.S. New Poly(Ionic Liquid) Based Fiber for Determination of Oxytetracycline in Milk Samples by Application of SPME-CE Technique. Molecules 2019, 24, 430.
- Moreno-González, D.; Hamed, A.M.; Gilbert-López, B.; Gámiz-Gracia, L.; García-Campaña, A.M. Evaluation of a Multiresidue Capillary Electrophoresis-Quadrupole-Time-of-Flight Mass Spectrometry Method for the Determination of Antibiotics in Milk Samples. J. Chromatogr. A 2017, 1510, 100–107.
- Díaz-Quiroz, C.A.; Francisco Hernández-Chávez, J.; Ulloa-Mercado, G.; Gortáres-Moroyoqui, P.; Martínez-Macías, R.; Meza-Escalante, E.; Serrano-Palacios, D. Simultaneous Quantification of Antibiotics in Wastewater from Pig Farms by Capillary Electrophoresis. J. Chromatogr. B 2018, 1092, 386–393.
- An, J.; Wang, X.; Ming, M.; Li, J.; Ye, N. Determination of Sulfonamides in Milk by Capillary Electrophoresis with [email protected] as a Dispersive Solid-Phase Extraction Sorbent. R. Soc. Open Sci. 2018, 5, 172104.
- Dai, T.; Duan, J.; Li, X.; Xu, X.; Shi, H.; Kang, W. Determination of Sulfonamide Residues in Food by Capillary Zone Electrophoresis with On-Line Chemiluminescence Detection Based on an Ag(III) Complex. Int. J. Mol. Sci. 2017, 18, 1286.
- Hong, Y.; Guo, X.; Chen, G.; Zhou, J.; Zou, X.; Liao, X.; Hou, T. Determination of Five Macrolide Antibiotic Residues in Milk by Micellar Electrokinetic Capillary Chromatography with Field Amplified Sample Stacking. J. Food Saf. 2018, 38.
- Lahouidak, S.; Soriano, M.L.; Salghi, R.; Zougagh, M.; Ríos, Á. Graphene Quantum Dots for Enhancement of Fluorimetric Detection Coupled to Capillary Electrophoresis for Detection of Ofloxacin. Electrophoresis 2019, 40, 2336–2341.
- Tejada-Casado, C.; Moreno-González, D.; Lara, F.J.; García-Campaña, A.M.; del Olmo-Iruela, M. Determination of Benzimidazoles in Meat Samples by Capillary Zone Electrophoresis Tandem Mass Spectrometry Following Dispersive Liquid–Liquid Microextraction. J. Chromatogr. A 2017, 1490, 212–219.
- Bosma, R.; Devasagayam, J.; Singh, A.; Collier, C.M. Microchip Capillary Electrophoresis Dairy Device Using Fluorescence Spectroscopy for Detection of Ciprofloxacin in Milk Samples. Sci. Rep. 2020, 10, 1–8.
- Tian, Z.; Gao, J.; Qin, W. Determination of Fluoroquinolones in Milk by Ionic Liquid-Mediated Two Phase Extraction Followed by Capillary Electrophoresis Analysis. Madr. J. Anal. Sci. Instrum. 2018, 3, 62–67.
- Kergaravat, S.V.; Nagel, O.G.; Althaus, R.L.; Hernández, S.R. Magneto Immunofluorescence Assay for Quinolone Detection in Bovine Milk. Food Anal. Methods 2020, 13, 1539–1547.
- Li, C.; Zhang, Y.; Eremin, S.A.; Yakup, O.; Yao, G.; Zhang, X. Detection of Kanamycin and Gentamicin Residues in Animal-Derived Food Using IgY Antibody Based Ic-ELISA and FPIA. Food Chem. 2017, 227, 48–54.
- Li, H.; Ma, S.; Zhang, X.; Li, C.; Dong, B.; Mujtaba, M.G.; Wei, Y.; Liang, X.; Yu, X.; Wen, K.; et al. Generic Hapten Synthesis, Broad-Specificity Monoclonal Antibodies Preparation, and Ultrasensitive ELISA for Five Antibacterial Synergists in Chicken and Milk. J. Agric. Food Chem. 2018, 66, 11170–11179.
- Wang, Z.; Hu, S.; Bao, H.; Xing, K.; Liu, J.; Xia, J.; Lai, W.H.; Peng, J. Immunochromatographic Assay Based on Time-Resolved Fluorescent Nanobeads for the Rapid Detection of Sulfamethazine in Egg, Honey, and Pork. J. Sci. Food Agric. 2020.
- Liang, X.; Sheng, Y.; Yu, W.; Zhao, S.; Shan, H.; Zhang, Q.; Wang, Z. Comparison of Chicken IgY and Mammalian IgG in Three Immunoassays for Detection of Sulfamethazine in Milk. Food Anal. Methods 2018, 11, 3452–3463.
- Shen, X.; Chen, J.; Lv, S.; Sun, X.; Dzantiev, B.B.; Eremin, S.A.; Zherdev, A.V.; Xu, J.; Sun, Y.; Lei, H. Fluorescence Polarization Immunoassay for Determination of Enrofloxacin in Pork Liver and Chicken. Molecules 2019, 24, 4462.
- Wang, G.; Xia, W.Q.; Liu, J.X.; Wang, J.P.; Liu, J. Directional Evolution of TetR Protein and Development of a Fluoroimmunoassay for Screening of Tetracyclines in Egg. Microchem. J. 2019, 150, 104184.
- Liu, H.; Sun, Y.; Jin, Z.; Yang, L.; Liu, J. Capillarity-Constructed Reversible Hot Spots for Molecular Trapping inside Silver Nanorod Arrays Light up Ultrahigh SERS Enhancement. Chem. Sci. 2013, 4, 3490–3496.
- Jiang, Y.; Sun, D.-W.; Pu, H.; Wei, Q. Surface Enhanced Raman Spectroscopy (SERS): A Novel Reliable Technique for Rapid Detection of Common Harmful Chemical Residues. Trends Food Sci. Technol. 2018, 75, 10–22.
- Wang, Q.; Zhao, W.-M. Optical Methods of Antibiotic Residues Detections: A Comprehensive Review. Sens. Actuators B Chem. 2018, 269, 238–256.
- Zhang, Y.; Teng, Y.; Qin, Y.; Ren, Z.; Wang, Z. Determination of Ciprofloxacin in Fish by Surface-Enhanced Raman Scattering Using a Liquid-Liquid Self-Assembled Gold Nanofilm. Anal. Lett. 2020, 53, 660–670.
- Jiang, X.; Qin, X.; Yin, D.; Gong, M.; Yang, L.; Zhao, B.; Ruan, W. Rapid Monitoring of Benzylpenicillin Sodium Using Raman and Surface Enhanced Raman Spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 140, 474–478.
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8.
- Mungroo, N.; Neethirajan, S. Biosensors for the Detection of Antibiotics in Poultry Industry—A Review. Biosensors 2014, 4, 472–493.
- Dixon, M.C. Quartz Crystal Microbalance with Dissipation Monitoring: Enabling Real-Time Characterization of Biological Materials and Their Interactions. J. Biomol. Tech. JBT 2008, 19, 151–158.
- Shen, Y.; Xu, L.; Li, Y. Biosensors for Rapid Detection of Salmonella in Food: A Review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 149–197.
- Tan, B.; Zhao, H.; Du, L.; Gan, X.; Quan, X. A Versatile Fluorescent Biosensor Based on Target-Responsive Graphene Oxide Hydrogel for Antibiotic Detection. Biosens. Bioelectron. 2016, 83, 267–273.
- Gaudin, V. Advances in Biosensor Development for the Screening of Antibiotic Residues in Food Products of Animal Origin-A Comprehensive Review. Biosens. Bioelectron. 2017, 90, 363–377.
- Wu, S.; Zhang, H.; Shi, Z.; Duan, N.; Fang, C.; Dai, S.; Wang, Z. Aptamer-Based Fluorescence Biosensor for Chloramphenicol Determination Using Upconversion Nanoparticles. Food Control 2015, 50, 597–604.
- Yue, F.; Li, F.; Kong, Q.; Guo, Y.; Sun, X. Recent Advances in Aptamer-Based Sensors for Aminoglycoside Antibiotics Detection and Their Applications. Sci. Total Environ. 2021, 762, 143129.




