
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Noemí García-Romero | + 1968 word(s) | 1968 | 2021-08-05 10:46:21 | | | |
| 2 | Amina Yu | Meta information modification | 1968 | 2021-09-02 05:40:10 | | |
Video Upload Options
Liquid biopsies are considered a good alternative and complementary tool for cancer management. The study of specific biomarkers by high throughput techniques could guide clinicians in the monitoring of disease evolution during the administration of targeted therapies. Although ddPCR has demonstrated its high sensitivity and specificity rates for detecting rare actionable mutations, further studies are required to implement it in all clinical laboratories for precision medicine.
1. Introduction
Despite many advances in the field, cancer remains one of the primary causes of death worldwide. In fact, estimations point to a total of 19.3 million new cases and almost 10 million deaths per year with no distinction between developed and undeveloped countries [1]. The most diagnosed cancer type worldwide is breast cancer followed by prostate cancer. On the other hand, lung cancer shows the highest mortality with an estimated 1,796,144 deaths worldwide in 2020 [2]. According to these statistics, it is not surprising that cancer has become the mainstay of research all over the world. In the last few years, many efforts in this field have been made, majorly in the diagnosis and prognosis of the different types of cancer, to assist clinicians to make more effective treatment selections through novel technologies. Nowadays, tumor tissue is the main source of information for diagnosis, stratification, prognosis, and treatment decision, discriminating between cancer or non-cancer lesions and providing significant information of different clinically relevant biomarkers [3][4][5].
Although tissue biopsies are crucial in the current evaluation and classification of cancer types, they present many limitations. Some of these relate to the highly invasive techniques required to obtain the biopsies and the risk to the patient associated with these procedures, as some tumors are hardly accessible due to their anatomical location and/or their infiltrative nature [5]. Also, tissue biopsies fail to represent the intra-tumoral heterogeneity since only a small fraction of the tumor obtained will be evaluated by physicians. To overcome this issue, liquid biopsies are present as a new minimally invasive tool for intra-tumoral monitoring which represents more accurately the tumoral status. Samples are collected from different biofluids, mostly from blood (serum and plasma) but also from saliva, breast milk, cerebrospinal fluid, stool, semen, urine, etc. [6]. Several biomarkers represented in Figure 1 could be isolated from different sources such as circulating tumor cells (CTCs), extracellular vesicles (EVs), cell-free DNA (cfDNA), and micro-RNA (miRNA).
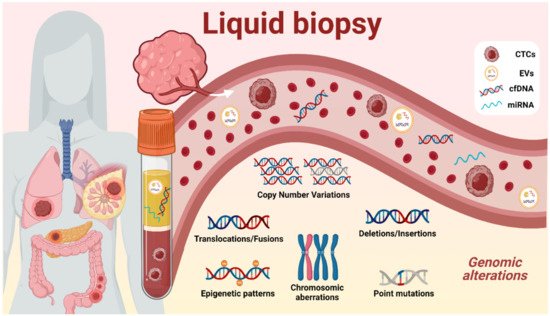
The circulating DNA found in all biofluids is referred to as cfDNA, which includes tumor-derived fraction called circulating tumor DNA (ctDNA) [7]. Nevertheless, cfDNA has been proven to be a powerful tool since all tumor cells, indistinctly of their phenotype, secrete DNA into the biofluids, providing information of the whole tumor, and revealing a snapshot of the intra-tumoral heterogeneity state at the moment of sample collection [8]. The mostly minimal-invasive nature of liquid biopsies allows clinicians to be informed about the molecular evolution of the tumor genetics, permitting disease monitoring, and avoiding the classical biopsies that could endanger the patient. The amount and length of ctDNA has been investigated due to its potential capacity for early detection and prognosis of some tumors. Also, in the current state of the art, ctDNA techniques are capable of detecting the genomic aberrations represented in Figure 1 , such as copy number variations (CNVs), methylation changes in DNA promoters, and single-nucleotide variations (SNVs) in a biofluidic sample [9], giving remarkable information about treatment response, tumoral staging, prognosis, minimal residual disease, and actionable mutations, enhancing more precise clinical decisions.
The near future of cancer diagnosis, prognosis, and treatment is expected to explore liquid biopsy biomarkers as an additional test to guide clinicians in every step of disease management. In this sense, an increasing number of cancer-derived biomarkers are being identified. In this review, we unify and classify different acknowledged and potentially actionable mutations that are relevant in lung, breast, colorectal, and pancreatic tumors, for further detection by ddPCR in clinical laboratories, in order to ease the way for clinicians in every step of disease management, striving to advance efforts in cancer diagnosis, prognosis, and treatment. To the best of our knowledge, this is the first report that presents a compendium of all the ongoing liquid biopsy-ddPCR clinical trials.
2. Breast Cancer
Hormonal therapies have greatly improved ER-positive mBC patient outcomes. Unfortunately, PIK3CA exon 9 E545K and E542K, and PIK3CA exon 20 H1047R, and H1047L mutations are frequently associated with resistance to hormonal therapies, such as fulvestrant [10][11]. For this reason, the identification of those biomarkers in plasma ctDNA could be used to predict BC treatment. In fact, a relationship has been shown between these mutations and a good response to anti-PI3K and anti-CDK4/6 targeted therapies together with fulvestrant in HR-positive HER2-negative advanced BC [12][13].
Another interesting gene to point out is the ESR1 ligand-binding domain (LBD) , which is mutated in 30–40% of ER-positive mBC patients [14][15]. Importantly, a significant number of mBC patients treated with first-line aromatase inhibitors (AIs) acquire ESR1 LBD mutations during the treatment developing endocrine treatment resistance [14][16][17]. Through ddPCR, several researchers have analyzed the ESR1 mutational status in peripheral blood [16][18][19]. The presence of ESR1 Y537S and D538G mutations in ER-positive mBC has been observed prior to treatment administration [20]. A significant increase in mutation prevalence was observed in patients who already received first-line AI therapy compared with those patients who only received adjuvant AI therapy [20]. Further analysis demonstrated the association of Y537S and D538G ESR1 mutations with worse OS.
The clinical significance of monitoring ESR1 LBD mutations Y537S, Y537N, Y537C, and D538G has been assessed in a cohort of sequential plasma samples from mBC and ER-positive primary BC treated with different endocrine therapies [21]. ddPCR data presented the ctDNA ESR1 mutation fluctuation as a consequence of treatment and showed that increasing amounts of ESR1 mutation post-therapy resulted in a poor response to treatment [21].
Even though ESR1 mutations have been principally detected in mBC, the ddPCR implementation allows detection of ESR1 mutations in approximately 2.5–7% of primary BC [22]. Furthermore, it has been observed that ESR1 mutations can be more frequently detected in cfDNA than in tissue biopsies [18].
3. Colon and Rectal Cancers
Over the last decade, most therapies have been designed to target aberrant signaling or activation of the MAPK pathway, such as the overexpression of EGFR found in 50–80% of patients [23][24][25]. Unfortunately, KRAS or BRAF mutations trigger non-responsiveness to anti- EGFR monoclonal antibody therapies [26][27][24]. KRAS mutations G12/G13 are observed in about 35–45% of CRC patients [28][24] and the first-line of treatment relies on a combination of chemotherapy of fluoropyrimidine with oxaliplatin or irinotecan [28]. BRAF shows lower mutation rates and appears in about 8–12% of metastatic CRC (mCRC), with V600E being the most common [29]. Although, the use of BRAF inhibitors alone such as vemurafenib did not reach the expected effectiveness, the combination with anti- EGFR monoclonal antibodies, MEK, and/or PI3K inhibitors have shown promising outcomes [29].
The DECALIB study was one of the first prospective studies to use the ddPCR technique to evaluate and compare the early detection of KRAS and BRAF mutations present in plasma cfDNA and tissue [30]. Afterwards, many other studies have also evaluated the concordance between tissue and plasma KRAS mutations obtaining elevated specificity and sensitivity rates [31][32]. Even though many different manuscripts have observed better correlations between cfDNA concentrations and the tumor mutation burden in mCRC [33][32][34], the cfDNA analysis could be also performed in patients at earlier cancer stages [31][32].
Longitudinal analysis of circulating KRAS concentrations in mCRC has great prognostic value since it has the ability of outcome prediction and treatment monitoring. In a KRAS -positive mCRC cohort from the prospective multicenter AIO KRK0207 trial (NCT00973609), ctDNA KRAS mutations were quantified via ddPCR before and 2–3 weeks after first-line chemotherapy initiation with fluoropyrimidine, oxaliplatin, and bevacizumab. Individuals with ctDNA KRAS mutations detected at baseline and in follow-up measurements presented worse OS and PFS. Remarkably, KRAS mutations identified at baseline in 15% of the patients were not detectable at follow-up measurements after treatment initiation. Those patients with KRAS mutation clearance at follow-up had better disease control and most notably better OS and PFS [28]. The same results have been observed by Holm et al. in patients included in the AXOAXI trial (NCT01531595 and EudraCT 2011-003137-33), treated with bevacizumab in combination with altering capecitabine and oxaliplatin or irinotecan [35], or by Kelin-Scory et al. where KRAS mutations cleared precipitously independently of type and intensity of chemotherapy and regardless of bevacizumab anti- VEGF treatment [36]. KRAS mutations have been detected 10 months earlier than radiographic confirmation of disease progression [23]. More remarkably, these ctDNA KRAS fluctuations and final disappearance open the possibility and potential treatment effectiveness of anti- EGFR therapies in those CRC patients [36][37].
Alternative sources of DNA such as EVs and fluids in the surrounding area of the tumoral tissue have been shown to provide information about disease evolution [38][39]. Fluids in the surrounding tumoral area such as the peritoneal fluid have been shown to be useful liquid biopsy sources of cfDNA [38]. In CRC patients with peritoneal metastases, a significantly higher amount of KRAS or BRAF ctDNA has been observed in peritoneal fluid than in plasma [40]. Since urine collection can be easily and repeatedly self-performed at any location with minimal effort, it has also been presented as an alternative source of cancer biomarkers for disease progression and drug response monitoring. In a proof-of-concept study with a KRAS -positive mCRC cohort, KRAS or BRAF mutations were screened from urine and matched plasma samples. Even though the concordance achieved was low, they showed the feasibility of using urine samples for non-urogenital tract tumor mutation screening [41].
4. Pancreatic Cancers
Although pancreatic cancer (PC), is not very frequent, its aggressiveness implies that the ratio of cases per number of deaths is close to 1. Nowadays, it is the seventh leading cause of cancer death worldwide and it has been estimated that it will surpass breast cancer in approximately 5 years [1]. Similar to CRC, the majority of patients are diagnosed with advanced stages and only 10–15% of PC patients have localized disease at the time of diagnosis [42]. Despite different genetic mutations identified in KRAS , CDKN2A , SMAD4, and TP53 genes, nearly all of them have failed to facilitate a treatment approach and patients continue receiving chemotherapy and radiotherapy depending on tumor stage [43][44].
Recently, the FDA approved olaparib, the first targeted therapy to increase PFS in metastatic PC patients with BRCA germline mutations [45].
As it has been assessed in different tumor types, bloodstream EVs represent an alternative source of ctDNA to provide information on disease evolution. In another study, ddPCR was used to assess KRAS hotspot mutations from EVs-derived DNA and matched cfDNA isolated from PC patients [46]. The sensitivity and specificity were 75.4% and 92.6%, respectively. Interestingly, KRAS mutation detection in EVs was superior to plasma cfDNA across all stages. Additionally, the observation of mutation rate in the localized pre- and post-resection cohort showed a precipitous decrease from 66% to 5%, respectively.
Recent studies confirmed the importance of genotyping different mutations in various genes aside from KRAS [47]. ddPCR was used to screen mutations previously identified via NGS in the KRAS , BRCA2 (S2378X), EGFR (R521K), ERBB2 (I655V, P1170A), and KRAS genes in a cohort of metastatic PDAC patients (NCT02017015). As expected, the KRAS mutation rate was 72.3%, whereas BRCA2 , EGFR, and ERBB2 were 11.7%, 13.3%, and 6.4%, respectively [48]. Other analyses associated the ERBB2 I655V mutation with worse OS among metastatic PC patients. Further analyses are required to determine whether those patients would benefit from targeted therapy using, for example, trastuzumab. These results remark the need for target gene sequencing analysis using ctDNA-based liquid biopsy samples to better guide individualized treatments.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 68, caac-21660.
- Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 28 June 2021).
- Lauby-Secretan, B.; Scoccianti, C.; Loomis, D.; Benbrahim-Tallaa, L.; Bouvard, V.; Bianchini, F.; Straif, K. Breast-Cancer Screening — Viewpoint of the IARC Working Group. N. Engl. J. Med. 2015, 372, 2353–2358.
- Lauby-Secretan, B.; Vilahur, N.; Bianchini, F.; Guha, N.; Straif, K. The IARC Perspective on Colorectal Cancer Screening. N. Engl. J. Med. 2018, 378, 1734–1740.
- Hüsing, A.; Kaaks, R. Risk prediction models versus simplified selection criteria to determine eligibility for lung cancer screening: An analysis of German federal-wide survey and incidence data. Eur. J. Epidemiol. 2020, 35, 899–912.
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548.
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. J. Exp. Clin. Cancer Res. 2020, 39, 120.
- Fernández-Lázaro, D.; García Hernández, J.L.; García, A.C.; Córdova Martínez, A.; Mielgo-Ayuso, J.; Cruz-Hernández, J.J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10, 215.
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10, 175883591879463.
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Prim. 2019, 5, 66.
- Jacot, W.; Dalenc, F.; Lopez-Crapez, E.; Chaltiel, L.; Durigova, A.; Gros, N.; Lozano, N.; Lacaze, J.-L.; Pouderoux, S.; Gladieff, L.; et al. PIK3CA mutations early persistence in cell-free tumor DNA as a negative prognostic factor in metastatic breast cancer patients treated with hormonal therapy. Breast Cancer Res. Treat. 2019, 177, 659–667.
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA -Mutated, Hormone Receptor–Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940.
- O’Leary, B.; Hrebien, S.; Morden, J.P.; Beaney, M.; Fribbens, C.; Huang, X.; Liu, Y.; Bartlett, C.H.; Koehler, M.; Cristofanilli, M.; et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 2018, 9, 896.
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Tomiguchi, M.; Sueta, A.; Murakami, K.; Omoto, Y.; Iwase, H. Analysis of ESR1 and PIK3CA mutations in plasma cell-free DNA from ER-positive breast cancer patients. Oncotarget 2017, 8, 52142–52155.
- Toy, W.; Shen, Y.; Won, H.; Green, B.; Sakr, R.A.; Will, M.; Li, Z.; Gala, K.; Fanning, S.; King, T.A.; et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013, 45, 1439–1445.
- Toy, W.; Weir, H.; Razavi, P.; Lawson, M.; Goeppert, A.U.; Mazzola, A.M.; Smith, A.; Wilson, J.; Morrow, C.; Wong, W.L.; et al. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discov. 2017, 7, 277–287.
- Guttery, D.S.; Page, K.; Hills, A.; Woodley, L.; Marchese, S.D.; Rghebi, B.; Hastings, R.K.; Luo, J.; Pringle, J.H.; Stebbing, J.; et al. Noninvasive Detection of Activating Estrogen Receptor 1 (ESR1) Mutations in Estrogen Receptor–Positive Metastatic Breast Cancer. Clin. Chem. 2015, 61, 974–982.
- Lupini, L.; Moretti, A.; Bassi, C.; Schirone, A.; Pedriali, M.; Querzoli, P.; Roncarati, R.; Frassoldati, A.; Negrini, M. High-sensitivity assay for monitoring ESR1 mutations in circulating cell-free DNA of breast cancer patients receiving endocrine therapy. Sci. Rep. 2018, 8, 4371.
- Urso, L.; Vernaci, G.; Carlet, J.; Lo Mele, M.; Fassan, M.; Zulato, E.; Faggioni, G.; Menichetti, A.; Di Liso, E.; Griguolo, G.; et al. ESR1 Gene Mutation in Hormone Receptor-Positive HER2-Negative Metastatic Breast Cancer Patients: Concordance Between Tumor Tissue and Circulating Tumor DNA Analysis. Front. Oncol. 2021, 11, 625636.
- Chandarlapaty, S.; Chen, D.; He, W.; Sung, P.; Samoila, A.; You, D.; Bhatt, T.; Patel, P.; Voi, M.; Gnant, M.; et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer. JAMA Oncol. 2016, 2, 1310.
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Inao, T.; Sueta, A.; Fujiwara, S.; Omoto, Y.; Iwase, H. Clinical significance of monitoring ESR1 mutations in circulating cell-free DNA in estrogen receptor positive breast cancer patients. Oncotarget 2016, 7, 32504–32518.
- Najim, O.; Huizing, M.; Papadimitriou, K.; Trinh, X.B.; Pauwels, P.; Goethals, S.; Zwaenepoel, K.; Peterson, K.; Weyler, J.; Altintas, S.; et al. The prevalence of estrogen receptor-1 mutation in advanced breast cancer: The estrogen receptor one study (EROS1). Cancer Treat. Res. Commun. 2019, 19, 100123.
- Misale, S.; Arena, S.; Lamba, S.; Siravegna, G.; Lallo, A.; Hobor, S.; Russo, M.; Buscarino, M.; Lazzari, L.; Sartore-Bianchi, A.; et al. Blockade of EGFR and MEK Intercepts Heterogeneous Mechanisms of Acquired Resistance to Anti-EGFR Therapies in Colorectal Cancer. Sci. Transl. Med. 2014, 6, 224ra26-224ra26.
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536.
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422.
- Lièvre, A.; Bachet, J.-B.; Boige, V.; Cayre, A.; Le Corre, D.; Buc, E.; Ychou, M.; Bouché, O.; Landi, B.; Louvet, C.; et al. KRAS Mutations As an Independent Prognostic Factor in Patients with Advanced Colorectal Cancer Treated with Cetuximab. J. Clin. Oncol. 2008, 26, 374–379.
- Amado, R.G.; Wolf, M.; Peeters, M.; Van Cutsem, E.; Siena, S.; Freeman, D.J.; Juan, T.; Sikorski, R.; Suggs, S.; Radinsky, R.; et al. Wild-Type KRAS Is Required for Panitumumab Efficacy in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 1626–1634.
- Lueong, S.S.; Herbst, A.; Liffers, S.-T.; Bielefeld, N.; Horn, P.A.; Tannapfel, A.; Reinacher-Schick, A.; Hinke, A.; Hegewisch-Becker, S.; Kolligs, F.T.; et al. Serial Circulating Tumor DNA Mutational Status in Patients with KRAS -Mutant Metastatic Colorectal Cancer from the Phase 3 AIO KRK0207 Trial. Clin. Chem. 2020, 66, 1510–1520.
- Sanz-Garcia, E.; Argiles, G.; Elez, E.; Tabernero, J. BRAF mutant colorectal cancer: Prognosis, treatment, and new perspectives. Ann. Oncol. 2017, 28, 2648–2657.
- Junca, A.; Tachon, G.; Evrard, C.; Villalva, C.; Frouin, E.; Karayan-Tapon, L.; Tougeron, D. Detection of Colorectal Cancer and Advanced Adenoma by Liquid Biopsy (Decalib Study): The ddPCR Challenge. Cancers 2020, 12, 1482.
- Zmrzljak, U.P.; Košir, R.; Krivokapić, Z.; Radojković, D.; Nikolić, A. Detection of Somatic Mutations with ddPCR from Liquid Biopsy of Colorectal Cancer Patients. Genes 2021, 12, 289.
- Liebs, S.; Keilholz, U.; Kehler, I.; Schweiger, C.; Haybäck, J.; Nonnenmacher, A. Detection of mutations in circulating cell-free DNA in relation to disease stage in colorectal cancer. Cancer Med. 2019, 8, 3761–3769.
- Galbiati, S.; Damin, F.; Burgio, V.; Brisci, A.; Soriani, N.; Belcastro, B.; Di Resta, C.; Gianni, L.; Chiari, M.; Ronzoni, M.; et al. Evaluation of three advanced methodologies, COLD-PCR, microarray and ddPCR, for identifying the mutational status by liquid biopsies in metastatic colorectal cancer patients. Clin. Chim. Acta 2019, 489, 136–143.
- Boysen, A.K.; Sørensen, B.S.; Lefevre, A.C.; Abrantes, R.; Johansen, J.S.; Jensen, B.V.; Schou, J.V.; Larsen, F.O.; Nielsen, D.; Taflin, H.; et al. Methodological development and biological observations of cell free DNA with a simple direct fluorescent assay in colorectal cancer. Clin. Chim. Acta 2018, 487, 107–111.
- Holm, M.; Andersson, E.; Osterlund, E.; Ovissi, A.; Soveri, L.-M.; Anttonen, A.-K.; Kytölä, S.; Aittomäki, K.; Osterlund, P.; Ristimäki, A. Detection of KRAS mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital PCR, Idylla, and next generation sequencing. PLoS ONE 2020, 15, e0239819.
- Klein-Scory, S.; Wahner, I.; Maslova, M.; Al-Sewaidi, Y.; Pohl, M.; Mika, T.; Ladigan, S.; Schroers, R.; Baraniskin, A. Evolution of RAS Mutational Status in Liquid Biopsies During First-Line Chemotherapy for Metastatic Colorectal Cancer. Front. Oncol. 2020, 10, 1–11.
- Raimondi, C.; Nicolazzo, C.; Belardinilli, F.; Loreni, F.; Gradilone, A.; Mahdavian, Y.; Gelibter, A.; Giannini, G.; Cortesi, E.; Gazzaniga, P. Transient Disappearance of RAS Mutant Clones in Plasma: A Counterintuitive Clinical Use of EGFR Inhibitors in RAS Mutant Metastatic Colorectal Cancer. Cancers 2019, 11, 42.
- Leick, K.M.; Kazarian, A.G.; Rajput, M.; Tomanek-Chalkley, A.; Miller, A.; Shrader, H.R.; McCarthy, A.; Coleman, K.L.; Kasi, P.M.; Chan, C.H.F. Peritoneal Cell-Free Tumor DNA as Biomarker for Peritoneal Surface Malignancies. Ann. Surg. Oncol. 2020, 27, 5065–5071.
- Yap, S.A.; Münster-Wandowski, A.; Nonnenmacher, A.; Keilholz, U.; Liebs, S. Analysis of cancer-related mutations in extracellular vesicles RNA by Droplet DigitalTM PCR. Biotechniques 2020, 69, 99–107.
- Van’t Erve, I.; Rovers, K.P.; Constantinides, A.; Bolhuis, K.; Wassenaar, E.C.E.; Lurvink, R.J.; Huysentruyt, C.J.; Snaebjornsson, P.; Boerma, D.; Broek, D.; et al. Detection of tumor-derived cell-free DNA from colorectal cancer peritoneal metastases in plasma and peritoneal fluid. J. Pathol. Clin. Res. 2021, 7, 203–208.
- Crisafulli, G.; Mussolin, B.; Cassingena, A.; Montone, M.; Bartolini, A.; Barault, L.; Martinetti, A.; Morano, F.; Pietrantonio, F.; Sartore-Bianchi, A.; et al. Whole exome sequencing analysis of urine trans-renal tumour DNA in metastatic colorectal cancer patients. ESMO Open 2019, 4, e000572.
- Sanabria Mateos, R.; Conlon, K.C. Pancreatic cancer. Surgery 2016, 34, 282–291.
- Takai, E.; Totoki, Y.; Nakamura, H.; Morizane, C.; Nara, S.; Hama, N.; Suzuki, M.; Furukawa, E.; Kato, M.; Hayashi, H.; et al. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci. Rep. 2015, 5, 1–10.
- Pelosi, E.; Castelli, G.; Testa, U. Pancreatic Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells. Biomedicines 2017, 5, 65.
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.-O.; Hochhauser, D.; Arnold, D.; Oh, D.-Y.; et al. Maintenance Olaparib for Germline BRCA -Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327.
- Allenson, K.; Castillo, J.; San Lucas, F.A.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747.
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Li, Q.K.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 1–6.
- Cheng, H.; Liu, C.; Jiang, J.; Luo, G.; Lu, Y.; Jin, K.; Guo, M.; Zhang, Z.; Xu, J.; Liu, L.; et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int. J. Cancer 2017, 140, 2344–2350.




