Through novel methodologies, including both basic and clinical research, progress has been made in the therapy of solid cancer. Recent innovations in anticancer therapies, including immune checkpoint inhibitor biologics, therapeutic vaccines, small drugs, and CAR-T cell injections, mark a new epoch in cancer research, already known for faster (epi-)genomics, transcriptomics, and proteomics. As the long-sought after personalization of cancer therapies comes to fruition, the need to evaluate all current therapeutic possibilities and select the best for each patient is of paramount importance. This is a novel task for medical care that deserves prominence in therapeutic considerations in the future. This is because cancer is a complex genetic disease. In its deadly form, metastatic cancer, it includes altered genes (and their regulators) that encode ten hallmarks of cancer-independent growth, dodging apoptosis, immortalization, multidrug resistance, neovascularization, invasiveness, genome instability, inflammation, deregulation of metabolism, and avoidance of destruction by the immune system. These factors have been known targets for many anticancer drugs and treatments, and their modulation is a therapeutic goal, with the hope of rendering solid cancer a chronic rather than deadly disease. In this article, the current therapeutic arsenal against cancers is reviewed with a focus on immunotherapies.
1. Introduction
Anticancer therapies aim to restrain the growth and spread of cancer. They can be divided in many ways. Traditionally, they are divided by the type of procedure, such as surgery, radiotherapy, chemotherapy, targeted therapy, immunotherapy, or cell therapy, but this distinction can sometimes be blurred because of combinatorial therapies, which are becoming increasingly attractive. It is the advent of immune checkpoint therapies that has alerted researchers to the possibility that cancer could soon become a chronic rather than a deadly disease. These therapies (which also include cell therapies like CAR-T cells) are currently being studied in over 3000 FDA-approved clinical studies (in phase 2 and 3) in various types of cancer as combinatorial therapies, with nearly all the tools that were previously available in the chemotherapy of cancer
[1]. We need to understand the essence of immune checkpoint immunotherapies if we wish to improve them by using small molecules as anticancer drugs in combination with them. Essentially, this is the aim of this mini review. I will summarize the current state of therapies, illustrate historical perspectives of approved anticancer treatments and explain strategies aimed at discovering novel anticancer drugs.
All treatments can be divided according to the characteristics of the cancer which they target and, in so doing, either inhibit or interfere with it. We lack knowledge of all molecular pathways, their genes and the epigenetic elements involved in malignant tissue transformation, its invasive spreading and metastasis. However, we know enough to begin to construct an idea about the genetic and epigenetic landscapes of each and every tumor type. There are worldwide projects aiming to map the cancer genetics and epigenetics as well as their proteomic landscape. Such data can be found in databases such as the Cancer Genomics, created by the National Cancer Institute (NCI) in 1997. This is an online reference on normal, pre-cancerous and cancerous genomes and, in addition, offers tools for viewing and analyzing the data, including browsing the genes reported for various phases of tumor progression. Seminal publications by Hanahan and Weinberg
[2][3] suggested that an eukaryotic cell can become malignant by accumulating at least one mutation in each of the ten characteristics (hallmarks) of cancer. We are now aware that cancer stem cells (CSC) can form by sequentially accumulating such changes over a long period of time (years), from which a cancer could develop. Importantly, although some are unique for each type of cancer, some changes can be common for many different cancer types. Hence, each of these cancer types might be treated by a combination therapy that targets common as well as specific cancer hallmarks.
2. Development of Cancer
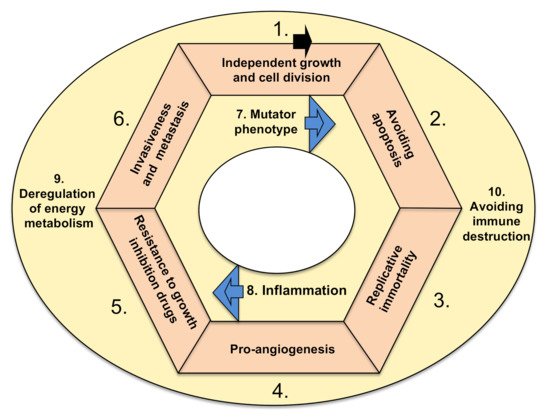
There are ten basic functional characteristics of cancer, which are called hallmarks. In order for a tumor to become malignant (i.e., able to invade surrounding tissues and metastasize) and lead to a deadly disease, either genetic or epigenetic changes must take place in all hallmarks (Figure 1). There are exceptions to this rule, which are rare. Namely, benign tumors (i.e., glioma, if in the head) or those localized inconveniently, preventing the life-supporting feature of the tissue, could be also deadly.
Figure 1. Hallmarks of cancer. The arrow denotes a possible order of new mutations occurring in cancer cells, forming a loop (modified, based on Hanahan and Weinberg
[2]). Tumor accumulates mutations (or epigenetic hits) and acquires listed hallmarks. Hallmarks 7 and 8 are accelerating features
[2]. Hallmarks 9 and 10 could occur anytime within the cycle, and I suggest that they are cancer-supporting characteristics.
The hallmarks are characterized by the following functional categories: (1) sustaining proliferative signaling, leading to independent growth, (2) resisting cell death (i.e., downregulation of apoptosis), (3) cellular immortalization (prolonging telomeres, enabling cell replication in perpetuity), (4) induction of angiogenesis, (5) resisting conventional therapies that include drugs as suppressors of growth, (6) invasive spreading with metastasis, (7) increased genetic/epigenetic instability (and attaining mutator phenotype), (8) tumor-promoting inflammation (as a growth-stimulating factor), (9) deregulation of energy metabolism in order to acquire more energy for growth (including metabolic shift from oxidative to anaerobic glycolysis) and (10) escaping destruction by the immune system
[2]. The ten hallmarks are depicted in
Figure 1. There is a hypothesis that various tissue types of cancer depend on particular combinations of genetic/epigenetic changes within the ten hallmarks. The drugs used in cancer therapies that can target and interfere with the ten hallmarks of cancer are elaborated below, firstly from the historic perspective of approval for therapy and secondly through their mode of action.
3. Cancer Stem Cells
Cancer is the expression used to describe an exceedingly complex group of diseases, caused by changes to the genes in one cell or a group of cells, resulting in altered protein–protein interactions. There are at least two major groups of hypotheses on how cancer can arise. One stipulates that it can arise from normal cells that undergo numerous gene mutations, epigenetic modifications, unrestrained signaling pathways and changes in the regulation of the microenvironment that follow the cycle of events depicted in Figure 1.
Alternatively, at some point during the changes within the genetic constellation of a particular cell in the affected tissue, a very small subpopulation of cancer cells can arise, called cancer stem cells, which then similarly follow a cycle of progression towards more malignant forms.
In the latter case, and in more detail, cancer forms a distinct tumor microenvironment, in which cancer stem cells (CSC) are tumor-(re)generating cells. Cancer stem cells exhibit self-renewal and multilineage capacities. CSCs divide, and a part of their progeny differentiates, while some CSCs mutate further, evolving into more invasive kinds of CSCs until they spread to the surrounding tissue and ultimately metastasize
[4][5]. Thus, in solid cancers, other stromal cells like cancer fibroblasts are derived in major part from cancer stem cell. However, part of the cancer includes normal cells that have migrated there, like tumor-infiltrating lymphocytes, macrophages, vascular epithelium or fibroblasts
[6]. Despite seeming normal, under the influence of cancer cells and the tumor microenvironment, cells from other parts of the body and outside the tumor mass begin to “behave” abnormally and perhaps might not have mutations in genes as CSCs in order to become a part of that particular cancer. Such could be the fate of many endothelial cells and fibroblasts, in general. We see also that lymphatic vessels can only be detected outside the tumor mass, probably because, inside solid tumors, they become “something else”. CSCs are considered resistant to chemotherapy or radiation, both of which can destroy other non-stem-cell tumor cells (and most cells from other parts of the body). CSCs are first described in myeloid leukemia
[7][8] and thereafter in solid tumors
[9][10]. CSCs’ roles were assessed in animal models
[11] and by studying their transcriptional profiles
[12]. CSCs resemble embryonic stem cells and induced pluripotent stem cells by expressing similar sets of transcription factors
[13][14]. It is thought that CSCs are made by the accumulation of genetic/epigenetic changes until they achieve the ability to self-renew, produce differentiated progeny and develop resistance to therapy. Alternatively, CSCs could be formed by reprogramming from a differentiated cell of a tissue at some point in the life of an individual
[15]. CSCs have a fundamental role in providing cancers with resistance to treatment, resurgence and metastasis. CSCs employ cell-signaling molecules in the Hedgehog, Wnt and Notch pathways, and drugs targeting these pathways showed encouraging results in experiments with multiple types of cancer
[16][17]. Therefore, targeting CSCs could be a promising strategy for anticancer therapy
[18].