
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Meilin Jin | + 5369 word(s) | 5369 | 2021-06-21 10:01:29 | | | |
| 2 | Conner Chen | Meta information modification | 5369 | 2021-07-01 04:22:14 | | | | |
| 3 | Conner Chen | Meta information modification | 5369 | 2021-07-01 04:23:34 | | |
Video Upload Options
Influenza is a highly known contagious viral infection that has been responsible for the death of many people in history with pandemics. These pandemics have been occurring every 10 to 30 years in the last century. The most recent global pandemic prior to COVID-19 was the 2009 influenza A (H1N1) pandemic. A decade ago, the H1N1 virus caused 12,500 deaths in just 19 months globally. Now, again, the world has been challenged with another pandemic. COVID-19 and influenza viruses have very similar signs, and symptoms may explain the similar origin. According to a recent World Health Organization survey, the COVID-19 attack and disease burden in children have been much lower than influenza outbreaks, and the secondary household attack rate has also been low. This is in stark contrast to reports of the virus spreading quickly in enclosed spaces like hospitals or cruise ships, as well as a high prevalence of healthcare-associated infections.
1. Virology and Structural Characteristics
1.1. Coronavirus (COVID-19)
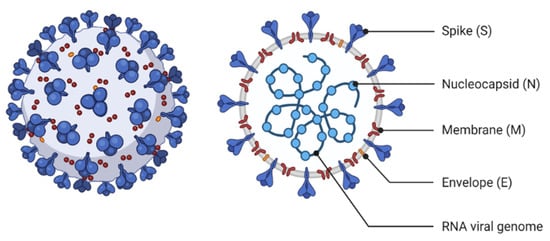
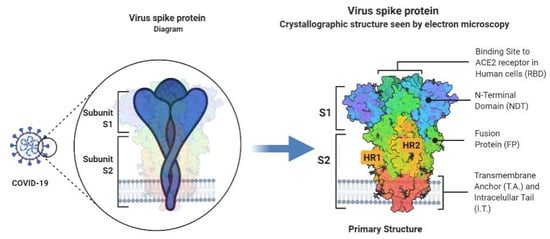
1.2. Influenza A (H1N1)
2. Epidemiology
2.1. Epidemiological Prospective of COVID-19
2.1.1. Death Rate by Age Group
| Gender proportion of COVID-19 cases | Age (Years) | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 |
| Male (%) | 47.5 | 50 | 52.8 | 52.3 | 51.8 | 53 | 49.7 | 39.2 | |
| Female (%) | 52.5 | 50 | 47.2 | 47.7 | 48.2 | 47 | 50.3 | 60.8 | |
| Gender proportion of COVID-19 deaths | Age (Years) | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 |
| Male (%) | 65 | 63.9 | 65.5 | 64.6 | 67.1 | 64.22 | 60.7 | 46.9 | |
| Female (%) | 35 | 36.1 | 34.5 | 35.4 | 32.9 | 35.8 | 39.3 | 53.1 | |
| Gender proportion of COVID-19 morbidity | Age (Years) | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 |
| Male (%) | 0.94 | 3.17 | 4.16 | 3.38 | 3.32 | 3.02 | 2.55 | 2.85 | |
| Female (%) | 1.08 | 3.22 | 3.67 | 2.93 | 2.84 | 2.36 | 2.04 | 2.56 |
2.1.2. Death Rate by Gender Ratio
2.1.3. Death Rate by Health Conditions
2.2. Epidemiological Prospective of Influenza A (H1N1)
2.2.1. Death Rate by Age Group
2.2.2. Death Rate by Sex Ratio
2.2.3. Death Rate by Health Conditions
3. Transmission and Replication
3.1. Transmission and Replication of COVID-19
3.1.1. Transmission
3.1.2. Replication
3.2. Transmission and Replication of Influenza A
3.2.1. Transmission
3.2.2. Replication
4. Clinical Manifestation
4.1. Clinical Manifestation of COVID-19
| Feature | COVID-19 | Influenza A(H1N1) | References | ||
|---|---|---|---|---|---|
| Epidemiology and Transmission | Fecal-oral transmission | Proved | Not proved | [39][50] | |
| Age composition | Most patients were older than 50 | Most patients were younger than 60 | |||
| Transmission mode | Asymptomatic/ symptomatic |
Symptomatic | [64] | ||
| Reproduction number | 3 | 1.5 | [52] | ||
| Incubation period | 4.9 | 1.4 | [51] | ||
| Treatment | Anti-viral drug | N3/ebselen/Remdesivir | Oseltamivir and zanamivir | [60][65][66] | |
| CP therapy | * | * | [67][68] | ||
| Vitamin D | * | * | [57][58] | ||
| MSC therapy | * | Not effective | [69] | ||
| Diagnosis | CRISPR-based SHERLOCK technique | * | Not develop | [70] | |
| qPCR | * | * | [71] | ||
| Gut microbiome | * | * | [72] | ||
| Lymphopenia | * | * | [73] | ||
| CT scan | Ground-glass opacities have frequently been placed in the periphery of lower lobes | Ground-glass opacities has a central, peripheral, or random distribution | [74] | ||
| Clinical manifestation | Acute lung injury | * | * | [44] | |
| Cardiovascular | * | * | [38] | ||
| Gastrointestinal | diarrhea | * | * | [75] | |
| nausea | * | * | |||
| vomiting | * | * | |||
| Molecular biology | Receptor for virus-cell entrance | ACE2 | Sialic acid receptor | [76][77] | |
| Genetic material | Just one positive-sense single-stranded RNA | Eight negative-sense single-stranded RNA | [78] | ||
| Location of replication | DMV (cytoplasm) | Nucleus | [19] | ||
4.1.1. Cardiovascular
4.1.2. Gastrointestinal (GI)
4.1.3. Lung
4.2. Clinical Manifestation of Influenza A (H1N1)
4.2.1. How Obesity Impacts Viral Infections?
4.2.2. Why Women Have Less Tendency to Be Affected by Viral Infections?
References
- Payne, S. Family Coronaviridae. Viruses 2017, 149–158.
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423.
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 2020, 78, 779–784.
- Satarker, S.; Nampoothiri, M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020, 51, 482–491.
- Mohammadi-Dehcheshmeh, M.; Moghbeli, S.M.; Rahimirad, S.; Alanazi, I.; Al Shehri, Z.S.; Ebrahimie, E. Achilles’ heel of SARS-CoV-2: Transcription regulatory sequence and leader sequence in 5’untranslated region have unique evolutionary patterns and are vital for virus replication in infected human cells. Preprints 2020.
- Christman, M.C.; Kedwaii, A.; Xu, J.; Donis, R.O.; Lu, G. Pandemic (H1N1) 2009 virus revisited: An evolutionary retrospective. Infect. Genet. Evol. 2011, 11, 803–811.
- Taubenberger, J.K.; Morens, D.M. The pathology of influenza virus infections. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 499–522.
- Fouchier, R.A.M.; Munster, V.; Wallensten, A.; Bestebroer, T.M.; Herfst, S.; Smith, D.; Rimmelzwaan, G.F.; Olsen, B.; Osterhaus, A.D.M.E. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 2005, 79, 2814–2822.
- Roxas, M.; Jurenka, J. Colds and influenza: A review of diagnosis and conventional, botanical, and nutritional considerations. Altern. Med. Rev. 2007, 12, 25–48.
- Hamilton, B.S.; Whittaker, G.R.; Daniel, S. Influenza virus-mediated membrane fusion: Determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses 2012, 4, 1144–1168.
- Arab-Mazar, Z.; Sah, R.; Rabaan, A.A.; Dhama, K.; Rodriguez-Morales, A.J. Mapping the incidence of the COVID-19 hotspot in Iran–Implications for Travellers. Travel Med. Infect. Dis. 2020, 34, 101630.
- Marson, F.A.L.; Ortega, M.M. COVID-19 in Brazil. Pulmonology 2020, 26, 241–244.
- Betancourt, W.Q.; Schmitz, B.W.; Innes, G.K.; Prasek, S.M.; Brown, K.M.P.; Stark, E.R.; Foster, A.R.; Sprissler, R.S.; Harris, D.T.; Sherchan, S.P.; et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021, 779, 146408.
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924.
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses—A statement of the Coronavirus Study Group. bioRxiv 2020.
- Verity, R.; Okell, L.C.; Dorigatti, I.; Winskill, P.; Whittaker, C.; Imai, N.; Cuomo-Dannenburg, G.; Thompson, H.; Walker, P.G.T.; Fu, H.; et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 2020, 20, 669–677.
- Undurraga, E.A.; Chowell, G.; Mizumoto, K. COVID-19 case fatality risk by age and gender in a high testing setting in Latin America: Chile, March–August 2020. Infect. Dis. Poverty 2021, 10, 11.
- Bhopal, S.S.; Bhopal, R. Sex differential in COVID-19 mortality varies markedly by age. Lancet 2020, 396, 532–533.
- Guilmoto, C.Z.Z. COVID-19 death rates by age and sex and the resulting mortality vulnerability of countries and regions in the world. medRxiv 2020.
- Scully, E.P.; Schumock, G.; Fu, M.; Massaccesi, G.; Muschelli, J.; Betz, J.; Klein, E.Y.; West, N.E.; Robinson, M.; Garibaldi, B.T.; et al. Sex and gender differences in COVID testing, hospital admission, presentation, and drivers of severe outcomes in the DC/Maryland region. medRxiv 2021.
- Takahashi, T.; Iwasaki, A. Sex differences in immune responses: Biological sex differences in immunity potentially underlie male bias for severe COVID-19. Science 2021, 371, 347–348.
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436.
- Moghadami, M.; Moattari, A.; Tabatabaee, H.R.; Mirahmadizadeh, A.; Rezaianzadeh, A.; Hasanzadeh, J.; Ebrahimi, M.; Zamiri, N.; Alborzi, A.; Bagheri Lankarani, K. High titers of hemagglutination inhibition antibodies against 2009 H1N1 influenza virus in Southern Iran. Iran. J. Immunol. 2010, 7, 39–48.
- Webster, R.G.; Kendal, A.P.; Gerhard, W. Analysis of antigenic drift in recently isolated influenza A (H1N1) viruses using monoclonal antibody preparations. Virology 1979, 96, 258–264.
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006, 12, 9–14.
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009, 459, 931–939.
- Balish, A.; Warnes, C.M.; Wu, K.; Barnes, N.; Emery, S.; Berman, L.; Shu, B.; Lindstrom, S.; Xu, X.; Uyeki, T.; et al. Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) Virus-United States, 2009. Morb. Mortal. Wkly. Rep. 2009, 58, 826–829.
- Lange, E.; Kalthoff, D.; Blohm, U.; Teifke, J.P.; Breithaupt, A.; Maresch, C.; Starick, E.; Fereidouni, S.; Hoffmann, B.; Mettenleiter, T.C.; et al. Pathogenesis and transmission of the novel swine-origin influenza virus A/H1N1 after experimental infection of pigs. J. Gen. Virol. 2009, 90, 2119–2123.
- Malveiro, D.; Flores, P.; Sousa, E.; Guimarães, J.C. The 2009 pandemic influenza A (H1N1) virus infection: Experience of a paediatric service at a third-level hospital in Lisbon, Portugal. Rev. Port. Pneumol. Engl. Ed. 2012, 18, 175–181.
- Al-Tawfiq, J.A.; Abed, M.; Saadeh, B.M.; Ghandour, J.; Shaltaf, M.; Babiker, M.M. Pandemic influenza A (2009 H1N1) in hospitalized patients in a Saudi Arabian hospital: Epidemiology and clinical comparison with H1N1-negative patients. J. Infect. Public Health 2011, 4, 228–234.
- Brockwell-Staats, C.; Webster, R.G.; Webby, R.J. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influenza Respir. Viruses 2009, 3, 207–213.
- Hillyard, R.D. Novel swine-origin influenza A (H1N1) virus investigation team emergence of a novel swime origin-inf A (H1N1) virus in humans. N. Engl. J. Med. 2009, 360, 2605–2615.
- Lemaitre, M.; Carrat, F. Comparative age distribution of influenza morbidity and mortality during seasonal influenza epidemics and the 2009 H1N1 pandemic. BMC Infect. Dis. 2010, 10, 162.
- Bhat, N.; Wright, J.G.; Broder, K.R.; Murray, E.L.; Greenberg, M.E.; Glover, M.J.; Likos, A.M.; Posey, D.L.; Klimov, A.; Lindstrom, S.E.; et al. Influenza-associated deaths among children in the United States, 2003–2004. N. Engl. J. Med. 2005, 353, 2559–2567.
- Dudley, J.P.; Mackay, I.M. Age-specific and sex-specific morbidity and mortality from avian influenza A (H7N9). J. Clin. Virol. 2013, 58, 568–570.
- Paskoff, T.; Sattenspiel, L. Sex-and age-based differences in mortality during the 1918 influenza pandemic on the island of Newfoundland. Am. J. Hum. Biol. 2019, 31, e23198.
- Noymer, A.; Garenne, M. The 1918 influenza epidemic’s effects on sex differentials in mortality in the United States. Popul. Dev. Rev. 2000, 26, 565–581.
- Rodríguez-Rieiro, C.; Carrasco-Garrido, P.; Hernández-Barrera, V.; de Andres, A.; Jimenez-Trujillo, I.; de Miguel, A.; Jiménez-García, R. Pandemic influenza hospitalization in Spain (2009): Incidence, in-hospital mortality, comorbidities and costs. Hum. Vaccines Immunother. 2012, 8, 443–447.
- Qian, Q.; Fan, L.; Liu, W.; Li, J.; Yue, J.; Wang, M.; Ke, X.; Yin, Y.; Chen, Q.; Jiang, C. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin. Infect. Dis. 2020.
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.-Y.; Chen, L.; Wang, M. Presumed asymptomatic carrier transmission of COVID-19. JAMA 2020, 323, 1406–1407.
- Chen, Y.; Wang, A.; Yi, B.; Ding, K.; Wang, H.; Wang, J.; Shi, H.; Wang, S.; Xu, G. The epidemiological characteristics of infection in close contacts of COVID-19 in Ningbo city. Chin. J. Epidemiol. 2020, 41, 668–672.
- Smith, J.C.; Sausville, E.L.; Girish, V.; Yuan, M.L.; Vasudevan, A.; John, K.M.; Sheltzer, J.M. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev. Cell 2020, 53, 514–529.
- de Celles, M.D.; Casalegno, J.-S.; Lina, B.; Opatowski, L. Influenza may facilitate the spread of SARS-CoV-2. medRxiv 2020.
- Linton, N.M.; Kobayashi, T.; Yang, Y.; Hayashi, K.; Akhmetzhanov, A.R.; Jung, S.; Yuan, B.; Kinoshita, R.; Nishiura, H. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: A statistical analysis of publicly available case data. J. Clin. Med. 2020, 9, 538.
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.-A.; Leibundgut, M.; Thiel, V.; Mühlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020, 27, 959–966.
- Asghari, A.; Naseri, M.; Safari, H.; Saboory, E.; Parsamanesh, N. The Novel Insight of SARS-CoV-2 Molecular Biology and Pathogenesis and Therapeutic Options. DNA Cell Biol. 2020, 39, 1741–1753.
- Zhu, X.; Wang, D.; Zhou, J.; Pan, T.; Chen, J.; Yang, Y.; Lv, M.; Ye, X.; Peng, G.; Fang, L.; et al. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 2017, 91, e00003-17.
- Li, Y.; Zhou, L.; Zhang, J.; Ge, X.; Zhou, R.; Zheng, H.; Geng, G.; Guo, X.; Yang, H. Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog. 2014, 10, e1004216.
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019, 10, 2342.
- Shu, T.; Huang, M.; Wu, D.; Ren, Y.; Zhang, X.; Han, Y.; Mu, J.; Wang, R.; Qiu, Y.; Zhang, D.-Y.; et al. SARS-Coronavirus-2 Nsp13 possesses NTPase and RNA helicase activities that can be inhibited by bismuth salts. Virol. Sin. 2020, 35, 321–329.
- Yuen, C.-K.; Lam, J.-Y.; Wong, W.-M.; Mak, L.-F.; Wang, X.; Chu, H.; Cai, J.-P.; Jin, D.-Y.; To, K.K.-W.; Chan, J.F.-W.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428.
- Wolff, G.; Limpens, R.W.A.L.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; de Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398.
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA proofreading: Molecular basis and therapeutic targeting. Mol. Cell 2020, 79, 710–727.
- Asadi, S.; ben Hnia, N.G.; Barre, R.S.; Wexler, A.S.; Ristenpart, W.D.; Bouvier, N.M. Influenza A virus is transmissible via aerosolized fomites. Nat. Commun. 2020, 11, 4062.
- Wu, Z.; Harrich, D.; Li, Z.; Hu, D.; Li, D. The unique features of SARS-CoV-2 transmission: Comparison with SARS-CoV, MERS-CoV and 2009 H1N1 pandemic influenza virus. Rev. Med. Virol. 2021, 31, e2171.
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 27, taaa021.
- Jiang, W.; Wang, Q.; Chen, S.; Gao, S.; Song, L.; Liu, P.; Huang, W. Influenza A virus NS1 induces G0/G1 cell cycle arrest by inhibiting the expression and activity of RhoA protein. J. Virol. 2013, 87, 3039–3052.
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A virus cell entry, replication, virion assembly and movement. Front. Immunol. 2018, 9, 1581.
- Liu, X.; Yang, C.; Hu, Y.; Lei, E.; Lin, X.; Zhao, L.; Zou, Z.; Zhang, A.; Zhou, H.; Chen, H.; et al. hisT1h1c regulates interferon-$β$ and inhibits influenza Virus replication by interacting with irF3. Front. Immunol. 2017, 8, 350.
- Liu, X.; Yang, C.; Sun, X.; Lin, X.; Zhao, L.; Chen, H.; Jin, M. Evidence for a novel mechanism of influenza A virus host adaptation modulated by PB 2-627. FEBS J. 2019, 286, 3389–3400.
- Song, L.; Liu, H.; Gao, S.; Jiang, W.; Huang, W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010, 84, 8849–8860.
- Shao, W.; Li, X.; Goraya, M.U.; Wang, S.; Chen, J.-L. Evolution of influenza a virus by mutation and re-assortment. Int. J. Mol. Sci. 2017, 18, 1650.
- Huang, S.S.H.; Banner, D.; Fang, Y.; Ng, D.C.K.; Kanagasabai, T.; Kelvin, D.J.; Kelvin, A.A. Comparative analyses of pandemic H1N1 and seasonal H1N1, H3N2, and influenza B infections depict distinct clinical pictures in ferrets. PLoS ONE 2011, 6, e27512.
- Bolsen, T.; Palm, R.; Kingsland, J.T. Framing the Origins of COVID-19. Sci. Commun. 2020, 42, 562–585.
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220.
- Zheng, Y.-Y.; Ma, Y.-T.; Zhang, J.-Y.; Xie, X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020, 17, 259–260.
- Leng, Z.; Zhu, R.; Hou, W.; Feng, Y.; Yang, Y.; Han, Q.; Shan, G.; Meng, F.; Du, D.; Wang, S.; et al. Transplantation of ACE2-mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020, 11, 216–228.
- Chaiwarith, R.; Prommee, N.; Liwsrisakun, C.; Oberdorfer, P.; Nuntachit, N.; Pothirat, C. A novel influenza A H1N1 clinical manifestations in patients at Chiang Mai University Hospital. J. Med. Assoc. Thail. 2011, 94, 908–915.
- Riquelme, A.; Alvarez-Lobos, M.; Pavez, C.; Hasbun, P.; Dabanch, J.; Cofre, C.; Jimenez, J.; Calvo, M. Gastrointestinal manifestations among Chilean patients infected with novel influenza A (H1N1) 2009 virus. Gut 2009, 58, 1567–1568.
- Fezeu, L.; Julia, C.; Henegar, A.; Bitu, J.; Hu, F.B.; Grobbee, D.E.; Kengne, A.-P.; Hercberg, S.; Czernichow, S. Obesity is associated with higher risk of intensive care unit admission and death in influenza A (H1N1) patients: A systematic review and meta-analysis. Obes. Rev. 2011, 12, 653–659.
- Kassir, R. Risk of COVID-19 for patients with obesity. Obes. Rev. 2020, 21, e13034.
- Long, C.; Xu, H.; Shen, Q.; Zhang, X.; Fan, B.; Wang, C.; Zeng, B.; Li, Z.; Li, X.; Li, H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020, 126, 108961.
- Dara, M.; Talebzadeh, M. CRISPR/Cas as a potential diagnosis technique for COVID-19. Avicenna J. Med. Biotechnol. 2020, 12, 201–202.
- Panning, M.; Eickmann, M.; Landt, O.; Monazahian, M.; Ölschläger, S.; Baumgarte, S.; Reischl, U.; Wenzel, J.J.; Niller, H.H.; Günther, S.; et al. Detection of influenza A (H1N1) v virus by real-time RT-PCR. Eurosurveillance 2009, 14, 19329.
- Huang, I.; Pranata, R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): Systematic review and meta-analysis. J. Intensive Care 2020, 8, 36.
- Cheng, Y.; Zhao, H.; Song, P.; Zhang, Z.; Chen, J.; Zhou, Y.-H. Dynamic changes of lymphocyte counts in adult patients with severe pandemic H1N1 influenza A. J. Infect. Public Health 2019, 12, 878–883.
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020, 71, 2669–2678.
- Deriu, E.; Boxx, G.M.; He, X.; Pan, C.; Benavidez, S.D.; Cen, L.; Rozengurt, N.; Shi, W.; Cheng, G. Influenza virus affects intestinal microbiota and secondary salmonella infection in the gut through type I interferons. PLoS Pathog. 2016, 12, e1005572.
- Smart, L.; Fawkes, N.; Goggin, P.; Pennick, G.; Rainsford, K.D.; Charlesworth, B.; Shah, N. A narrative review of the potential pharmacological influence and safety of ibuprofen on coronavirus disease 19 (COVID-19), ACE2, and the immune system: A dichotomy of expectation and reality. Inflammopharmacology 2020, 28, 1141–1152.
- Deng, Q.; Hu, B.; Zhang, Y.; Wang, H.; Zhou, X.; Hu, W.; Cheng, Y.; Yan, J.; Ping, H.; Zhou, Q. Suspected myocardial injury in patients with COVID-19: Evidence from front-line clinical observation in Wuhan, China. Int. J. Cardiol. 2020, 311, 116–121.
- Zhong, P.; Xu, J.; Yang, D.; Shen, Y.; Wang, L.; Feng, Y.; Du, C.; Song, Y.; Wu, C.; Hu, X.; et al. COVID-19-associated gastrointestinal and liver injury: Clinical features and potential mechanisms. Signal Transduct. Target. Ther. 2020, 5, 256.
- Aziz, M.; Haghbin, H.; Lee-Smith, W.; Goyal, H.; Nawras, A.; Adler, D.G. Gastrointestinal predictors of severe COVID-19: Systematic review and meta-analysis. Ann. Gastroenterol. 2020, 33, 615–630.
- Leng, L.; Cao, R.; Ma, J.; Mou, D.; Zhu, Y.; Li, W.; Lv, L.; Gao, D.; Zhang, S.; Gong, F.; et al. Pathological features of COVID-19-associated lung injury: A preliminary proteomics report based on clinical samples. Signal Transduct. Target. Ther. 2020, 5, 240.
- Puzelli, S.; Buonaguro, F.M.; Facchini, M.; Palmieri, A.; Calzoletti, L.; De Marco, M.A.; Arace, P.; De Campora, E.; Esposito, C.; Cassone, A.; et al. Cardiac tamponade and heart failure due to myopericarditis as a presentation of infection with the pandemic H1N1 2009 influenza A virus. J. Clin. Microbiol. 2010, 48, 2298–2300.
- Maines, T.R.; Jayaraman, A.; Belser, J.A.; Wadford, D.A.; Pappas, C.; Zeng, H.; Gustin, K.M.; Pearce, M.B.; Viswanathan, K.; Shriver, Z.H.; et al. Transmission and pathogenesis of swine-origin 2009 A (H1N1) influenza viruses in ferrets and mice. Science 2009, 325, 484–487.
- Gan, H.; Hao, Q.; Idell, S.; Tang, H. Transcription factor Runx3 is induced by influenza A virus and double-strand RNA and mediates airway epithelial cell apoptosis. Sci. Rep. 2015, 5, 17916.
- de Siqueira, J.V.V.; Almeida, L.G.; Zica, B.O.; Brum, I.B.; Barceló, A.; de Siqueira Galil, A.G. Impact of obesity on hospitalizations and mortality, due to COVID-19: A systematic review. Obes. Res. Clin. Pract. 2020, 14, 398–403.
- Conti, P.; Younes, A. Coronavirus COV-19/SARS-CoV-2 affects women less than men: Clinical response to viral infection. J. Biol. Regul. Homeost. Agents 2020, 34, 339–343.
- Sama, I.E.; Ravera, A.; Santema, B.T.; van Goor, H.; ter Maaten, J.M.; Cleland, J.G.F.; Rienstra, M.; Friedrich, A.W.; Samani, N.J.; Ng, L.L.; et al. Circulating plasma concentrations of ACE2 in men and women with heart failure and effects of renin-angiotensin-aldosterone-inhibitors. Eur. Heart J. 2020, 41, 1810–1817.




