
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mohamed Shehata | + 1133 word(s) | 1133 | 2021-04-14 08:23:17 | | | |
| 2 | Karina Chen | Meta information modification | 1133 | 2021-04-15 02:45:29 | | |
Video Upload Options
Histopathology is a significant branch of biology that covers the investigation of the cell anatomy and tissues of organisms at a microscopic level by a histopathologist. Histopathological images are very influential for the final decision procedure of effective therapeutics; these images are essential to investigate the status of a certain biological structure and to diagnose diseases like cancer . Digital histopathology represents a significant evolution in modern medicine. It often uses machine vision techniques as a basis. Nevertheless, because of the special properties of digital histopathology images and their processing tasks, specific processing approaches are usually needed.
1. Histopathology Images Background
Uropathologists use different screening methods to determine the various tumor histology in the prostate in a good quality. Typical tissue of prostate incorporates glands and stroma. The gland is the basic anatomical structural unit of the prostate. The stroma is the fibromuscular tissue around glands [1]. Each gland unit consists of a lumen and rows of epithelial layers surrounding the lumen. The stroma keeps the gland units together. When cancer is in high-grade, stroma and lumen are both replaced by epithelial cells [2]. Once the slides are stained using a hematoxylin and eosin (H&E) solution, the nuclei become dark blue and the epithelial layer and stroma become several shades of purple to pink [1].
To date, one of the most effective ways to measure aggressiveness of prostate cancer is using the Gleason grading system [2][3][4]. The Gleason grading system is completely founded on architectural arrangements of prostatic carcinoma, and a substantial parameter to a therapeutic final decision. Gleason grading has five grade groups from grade 1 (G1) to grade 5 (G5), with a grade of G1 refers to tissue with a maximum grade of resemblance to normal tissue and outstanding prognosis, and a grade of G5 refers to poorly differentiated tissue and the worst prediction [2][5]. Artificial intelligence has the ability to improve the quality of Gleason grading. Abundant automated Gleason grading systems were proposed and have led to increased consistency [6][5][7][8][9][10][11][12].
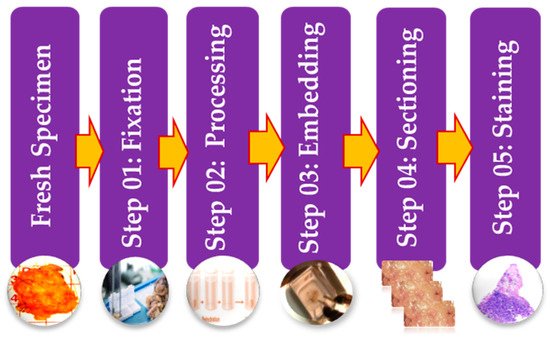
Histopathology images can be acquired by using specialized cameras with a microscope wherein an automated computerized approach can be carried out [13]. To study various architecture and constituent of tissues under a microscope, the biopsy specimen is embedded in wax and dyed with one or more stains. Staining procedures are used by pathologists for cellular components separation for structural in addition to component visualization of tissue for diagnosis [14]. Stages of the preparation process of the tissue slides are as presented in Figure 3. It consists of five operations, and each of them can affect the quality of the final image [14][15]. (I) Fixation: Samples of biological tissues are fixed with chemical fixation. There are many ways of fixation, but the commonly applied way in the biomedical field is fixation with formaldehyde or glutaraldehyde solution to protect the cells [12]. This is a critical step in tissue preparation and aims to prevent tissue autolysis and putrefaction; (II) Processing: Tissue processing is a crucial step and involves two main processes: dehydration and clearing. Dehydration is used to extract water from the gross tissue and substitute it with a certain concentration of alcohol which solidifies it [16]. This process helps incise superfine sections of the specimen. Clearing consists of removing the dehydrator with a material that will be the solvent in both the embedding paraffin and the dehydrating agent; (III) Tissue Embedding: Thus is the process wherein tissues are carefully positioned in a medium such as wax [12], so when solidified, it will provide enough external support to allow very thin sectioning. This process is essential as the proper tissue orientation is necessary for precise microscopic evaluation; (IV) Sectioning: this process is required to generate superfine slices of tissue samples sufficient such that the details of the microstructure characterization of the cells can be obviously noticed using microscopy methods. After that, carry the superfine slices of sample onto a clean glass slide [14]; (V) Staining: The final step in preparing tissue for light microscopy is to stain it and mount it on the slide. Staining increases contrast to the tissue and, also highlights some specific features which would otherwise be practically invisible in the microscope [14]. There are many types of stain but the most common type of staining for histology is H & E.
Automated prostate cancer diagnosis using histopathology images is deemed to offer great promise for advanced cancer therapy, however, it is not a simple task, as several open scientific challenges have to be overcome before the CAD system of histopathology images can become part of the routine healthcare diagnostic pipeline. These challenges occur because of the numerous technical and computational variabilities and artifacts incurred due to differences in slide preparation and because of the complicated structure of the tumor tissues architecture [17]. Image analysis techniques are substantially reliant on the quality of the digital slide images.
3. Histopathology Image Analysis Methodology
Digitized histopathology is a current direction that makes huge numbers of images available for automated analysis. It enables visualization and interpretation of pathology cells and tissue samples in a great resolution images and with the assistance of software tools [18][19]. This opens a new era to design image analysis techniques that assist clinicians and promote their image descriptions (e.g., grading, staging) with the purpose of image features quantification. In that respect, the computer-aided diagnosis of histological image analysis is a newly challenging domain for biomedical image analysis. CAD can be defined as detecting cancer within the examined tissue using computer software [20][21][22], which is the main mission of the pathologist [23]. The combination of conventional diagnosis techniques with computational AI techniques provides a good possibility to decrease the workload of pathologists while preserving performance. There is a need for a precise CAD system that minimizes reading interpretation times, lowers necessary experience in anatomic pathology, and provides a consistent risk evaluation of cancer existence in prostate histopathology images without additional burden to pathologists. Such a CAD system would automatically find out suspected lesions in prostate histopathology images to assist screen for prostate cancer in large patient populations. A typical CAD system for detecting prostate cancer receives raw histopathological images, preprocesses them, and produces a particular diagnostic result [24].
Over the last two decades, numerous research papers on CAD systems were published. Automated systems for digital histopathological imaging can maintain reproducibility and consistency using suitable image processing techniques [17]. In fact, there are many research perspectives for CAD systems applied in the histopathological domain, including: (I) cancer detection in the given tissue, (II) automatic grading to correctly quantify the level of the malignancy, which can offer more insights into disease characterization, (III) cell/nuclei/gland segmentation that discovers and separates these regions from images, and (IV) multi-class classification for the different subtypes of a specific type of cancer.
References
- Rashid, S.; Fazli, L.; Boag, A.; Siemens, R.; Abolmaesumi, P.; Salcudean, S.E. Separation of Benign and Malignant Glands in Prostatic Adenocarcinoma. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nagoya, Japan, 22–26 September 2013; Springer International Publishing: Berlin/Heidelberg, Germany, 2013; pp. 461–468.
- Li, W.; Li, J.; Sarma, K.V.; Ho, K.C.; Shen, S.; Knudsen, B.S.; Gertych, A.; Arnold, C.W. Path R-CNN for Prostate Cancer Diagnosis and Gleason Grading of Histological Images. IEEE Trans. Med. Imaging 2019, 38, 945–954.
- Humphrey, P.A. Histopathology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a030411.
- Van Booven, D.J.; Kuchakulla, M.; Pai, R.; Frech, F.S.; Ramasahayam, R.; Reddy, P.; Parmar, M.; Ramasamy, R.; Arora, H. A Systematic Review of Artificial Intelligence in Prostate Cancer. Res. Rep. Urol. 2021, 13, 31–39.
- Bulten, W.; Pinckaers, H.; van Boven, H.; Vink, R.; de Bel, T.; van Ginneken, B.; van der Laak, J.; de Kaa, C.H.; Litjens, G. Automated gleason grading of prostate biopsies using deep learning. arXiv 2019, arXiv:1907.07980. Available online: (accessed on 10 January 2021).
- Arvaniti, E.; Fricker, K.S.; Moret, M.; Rupp, N.; Hermanns, T.; Fankhauser, C.; Wey, N.; Wild, P.J.; Rüschoff, J.H.; Claassen, M. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci. Rep. 2018, 8, 1–11.
- Kallen, H.; Molin, J.; Heyden, A.; Lundstrom, C.; Astrom, K. Towards grading gleason score using generically trained deep convolutional neural networks. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1163–1167.
- Nir, G.; Hor, S.; Karimi, D.; Fazli, L.; Skinnider, B.F.; Tavassoli, P.; Turbin, D.; Villamil, C.F.; Wang, G.; Wilson, R.S.; et al. Automatic grading of prostate cancer in digitized histopathology images: Learning from multiple experts. Med. Image Anal. 2018, 50, 167–180.
- Doyle, S.; Hwang, M.; Shah, K.; Madabhushi, A.; Feldman, M.; Tomaszeweski, J. Automated grading of prostate cancer using architectural and textural image features. In Proceedings of the 2007 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Arlington, VA, USA, 12–15 April 2007; pp. 1284–1287.
- Niazi, M.K.K.; Yao, K.; Zynger, D.L.; Clinton, S.K.; Chen, J.; Koyuturk, M.; LaFramboise, T.; Gurcan, M. Visually Meaningful Histopathological Features for Automatic Grading of Prostate Cancer. IEEE J. Biomed. Health Inform. 2017, 21, 1027–1038.
- Khurd, P.; Bahlmann, C.; Maday, P.; Kamen, A.; Gibbs-Strauss, S.; Genega, E.M.; Frangioni, J.V. Computer-aided Gleason grading of prostate cancer histopathological images using texton forests. In Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, The Netherlands, 14–17 April 2010; pp. 636–639.
- Slaoui, M.; Fiette, L. Histopathology Procedures: From Tissue Sampling to Histopathological Evaluation. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2010; Volume 691, pp. 69–82.
- Aswathy, M.; Jagannath, M. Detection of breast cancer on digital histopathology images: Present status and future possibilities. Inform. Med. Unlocked 2017, 8, 74–79.
- Belsare, A. Histopathological Image Analysis Using Image Processing Techniques: An Overview. Signal Image Process. Int. J. 2012, 3, 23–36.
- Li, C.; Chen, H.; Li, X.; Xu, N.; Hu, Z.; Xue, D.; Qi, S.; Ma, H.; Zhang, L.; Sun, H. A review for cervical histopathology image analysis using machine vision approaches. Artif. Intell. Rev. 2020, 53, 4821–4862.
- Cahill, L.C.; Fujimoto, J.G.; Giacomelli, M.G.; Yoshitake, T.; Wu, Y.; Lin, D.I.; Ye, H.; Carrasco-Zevallos, O.M.; Wagner, A.A.; Rosen, S. Comparing histologic evaluation of prostate tissue using nonlinear microscopy and paraffin H&E: A pilot study. Mod. Pathol. 2019, 32, 1158–1167.
- Das, A.; Nair, M.S.; Peter, S.D. Computer-Aided Histopathological Image Analysis Techniques for Automated Nuclear Atypia Scoring of Breast Cancer: A Review. J. Digit. Imaging 2020, 33, 1–31.
- Jimenez-del-Toro, O.; Otálora, S.; Andersson, M.; Eurén, K.; Hedlund, M.; Rousson, M.; Atzori, M. Analysis of histopathology images: From traditional machine learning to deep learning. In Biomedical Texture Analysis; Academic Press: Cambridge, MA, USA, 2017; pp. 281–314.
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175.
- Veta, M.M.; Pluim, J.P.W.; Van Diest, P.J.; Viergever, M.A. Breast Cancer Histopathology Image Analysis: A Review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411.
- McClure, P.; Elnakib, A.; El-Ghar, M.A.; Khalifa, F.; Soliman, A.; El-Diasty, T.; Suri, J.S.; Elmaghraby, A.; El-Baz, A. In-Vitro and In-Vivo Diagnostic Techniques for Prostate Cancer: A Review. J. Biomed. Nanotechnol. 2014, 10, 2747–2777.
- Reda, I.; Khalil, A.; Elmogy, M.; El-Fetouh, A.A.; Shalaby, A.; El-Ghar, M.A.; Elmaghraby, A.; Ghazal, M.; El-Baz, A. Deep Learning Role in Early Diagnosis of Prostate Cancer. Technol. Cancer Res. Treat. 2018, 17, 1533034618775530.
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42.
- Wang, S.; Burtt, K.; Turkbey, B.; Choyke, P.; Summers, R.M. Computer Aided-Diagnosis of Prostate Cancer on Multiparametric MRI: A Technical Review of Current Research. BioMed Res. Int. 2014, 2014, 1–11.





