1000/1000
Hot
Most Recent

Proanthocyanidins are essential secondary plant metabolites that contribute to the nutritional value and sensory quality of many fruits and the related processed products. The accumulation of proanthocyanidins is associated with the resistance of plants against a broad spectrum of abiotic and biotic stress conditions. Many specific regulators were involved in the proanthocyanidins biosynthetic network in response to various environmental conditions.
Horticultural crops, including vegetables and fruits, are a crucial food source for human nutrition as they contain carbohydrates, proteins, vitamins, organic acids, and minerals. Proanthocyanidins (PAs), also known as condensed tannins, are among the most abundant polyphenols in plants. They are naturally present in woody plants and herbaceous species. PAs are present in the leaves, fruits, seeds, roots, and other parts of plants[1], and play essential roles in the growth and development of leaves and fruits, as well as, the modulation of seed dormancy and germination[2][3][4][5][6][7]. In addition, PAs also influence mouthfeel by enhancing the astringency of fruits and in their processed products, e.g., wines and beverages[8][9]. Moreover, as potential dietary antioxidants, PAs are considered beneficial to human health. It has been widely accepted that the PAs from food plants and medicinal plants are associated with various bioactivities, displaying immunomodulatory, anti-inflammatory, anti-cancer, anti-microbial, and hypolipidemic properties while reducing the risk of cardiovascular disease and ameliorating obesity [10][11][12][13][14][15][16][17][18][19][20][21][22].
Developmental regulation is considered the driving factor for PAs biosynthesis, which can also be influenced by environmental fluctuations. In grape berry, the PAs accumulate in the skin prior to véraison, after which the total PAs content decreases due to intermediate metabolites deviation[4]. Many crops have shown a similar decreasing trend in PAs levels during the fruit ripening process, including bilberries, persimmons, and blueberries[23][24][25]. PAs contribute to the astringency and bitterness of the young fruits, preventing them from being eaten before they are ripe[26]. The accumulation of PAs is associated with the resistance of plants against various biotic and abiotic stimuli, such as low temperature[27][28], drought[29], wounding[30][31], UV radiation[30], and fungal pathogens[32][33][34]. Therefore, increasing attention has been focused on the metabolic engineering and regulatory mechanism of PAs in horticultural plants. The structural genes and key factors that regulate PAs biosynthesis were identified and characterized in many plants, particularly in horticultural crops rich in PAs, such as grapevines, poplars, and apples. Studies involving PAs biosynthesis regulation in response to environmental stresses revealed that the regulatory network was based on the regulators belonging to the MYB, MYC-like basic helix-loop-helix (bHLH), WD40-repeat proteins, and the MYB-bHLH-WD40 (MBW) complexes. However, research regarding more specific regulators in this complicated network is required to understand the interaction between these vital regulators during PAs biosynthesis to ultimately improve the quality of crops.
PAs consist of a mixture of flavan-3-ol units and flavan-3,4-diols (leucoanthocyanidins) in complicated ways[35]. Flavan-3-ol monomeric units include (+)-catechins, (−)-epicatechins, and their derivatives: (+)-gallocatechin, (+)-catechin gallate, (+)-gallocatechin gallate (−)-epicatechin gallate, (−)-epigallocatechin, and (−)-epigallocatechin gallate (Figure 1). Both (+)-catechins and (−)-epicatechins are believed to act as the starter units for PAs polymerization, and flavan-3-ols and flavan-3,4-diols serve as extension units for further PAs extension[1][36][37]. Different numbers of subunits polymerized to form oligomeric or polymeric PAs[1][38]. The degree of polymerization of PAs varies among species and tissues. Based on the interflavanic linkages, the subunits linked by C4-C8 and/or C4-C6 bonds are known as B-type PAs, and those with additional C2-O-C7 or C2-O-C5 bond in the structure are classified into A-type PAs (Figure 1)[1]. In addition, esterification, glycosylation, and other modifications have also been found in PAs with more complex structures. B-type PAs are widely distributed in plants and foods, such as senna alata leaves, pine trees, black wattle, beans, barley, sorghum, seeds, fruits, berries, nuts, cinnamon, chocolate, and wine[39][40][41][42][43][44][45]. A-type PAs are mainly found in bilberry, peanuts, plums, cranberries, curry, and cinnamon[46][47][48][49].
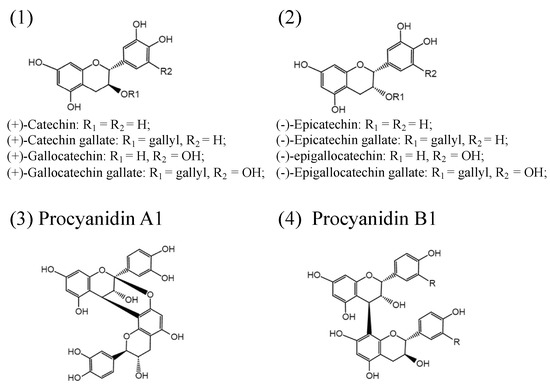
Figure 1. Structures of the flavan-3-ols (1,2); simple A1-type proanthocyanidins (3); simple B1-type proanthocyanidins (4).
The pathway of flavonoid biosynthesis has been well studied in various plants (Figure 2). Flavonols, anthocyanins, and PAs are the main products of the flavonoid biosynthetic pathway, which belongs to the plant secondary metabolism[50][51][52]. They share the common upstream steps in the biosynthetic pathway of flavonoids, and their precursor is dihydroflavonols which can be converted to leucoanthocyanidins by dihydroflavonol-4-reductase (DFR) and to flavonols by flavonol synthase (FLS). Leucoanthocyanidins can be further catalyzed to produce anthocyanidins by the oxygenation reaction of anthocyanidin synthase (ANS) or leucoanthocyanidin dioxygenase (LDOX). Leucoanthocyanidins and anthocyanidins can be catalyzed by leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) to produce 2,3-trans-flavan-3-ols (such as (+)-catechin) and 2,3-cis-flavan-3-ols (such as (−)-epicatechin) which are the key subunits involved in the last steps of PAs biosynthesis.
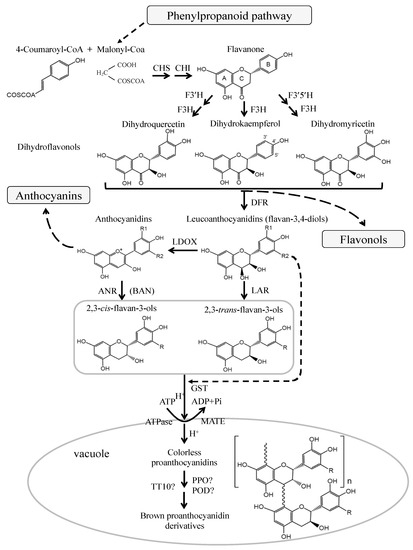
Figure 2. Biosynthetic pathway of flavonoid (adapt from[1][38]). The enzymes are: CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; DFR, dihydroflavonol-4-reductase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase; LDOX, leucoanthocyanidin dioxygenase; GST, glutathione S-transferase; MATE, multidrug and toxic compound extrusion; PPO, polyphenol oxidase; POD, peroxidase; TT10, transparent testa 10.
To date, LAR and ANR genes have been cloned from several plant species, and their functions have been demonstrated both genetically and biochemically[3][36][53][54][55][56][57][58][59][60]. As is well known, leucoanthocyanidins are converted to (+)-catechins through the catalysis of LAR. Interestingly, ectopic expression of tea CsLAR, and cacao TcLAR in tobacco showed that the production of (−)-epicatechin was higher than that of catechins[57][61]. More recent studies also found that LAR from Medicago truncatula and grapevine can convert 4β-(S-cysteinyl)-epicatechin into (−)-epicatechins, thereby regulating the degree of PAs polymerization[59][62]. In addition to catalyzing the synthesis of (−)-epicatechins from anthocyanidins, ANR is proven to convert anthocyanidins to a mixture of (−)-epicatechins and (+)-catechins[61][63]. This suggests that LAR and ANR may possess more features to be explored [61][62][63].
Most of the enzymes for flavonoid biosynthesis are located on the endoplasmic reticulum and tonoplast of cells. PAs are synthesized in the cytoplasm, and then transferred and accumulated into the vacuole and apoplast [64][65][66]. In seed, PAs are mostly accumulated in the inner endothelial layer of seed testa [35][67][36], and the brown color of the seed coat may result from the oxidation of PAs[68]. The seeds without PAs content are called transparent testa (tt) phenotype and tannin deficient seeds, which exhibit yellow or pale brown in color[54][69][70]. The PA-deficient mutants have become an ideal material for investigating the biosynthesis and transportation of PAs. MATE (multidrug and toxic compound extrusion) and GST (glutathione S-transferase) are proteins responsible for flavonoids transportation. AtTT12, a member of the MATE family, localizes the PAs in the vacuoles of the testa of Arabidopsis seeds[71]. AtTT13/AtAHA10 encodes a protein belonging to the ATPase family, which is required for the deposition of PA precursors in vacuole and functions as a proton pump, providing the driving force for TT12-mediated transportation of PA precursors[72]. VvMATE1 and VvMATE2 are transport proteins located in the tonoplast and Golgi complex, respectively, and involved in the transport and accumulation of PAs through different ways in grapevine [66]. GST proteins were thought to deliver anthocyanins into vacuoles, e.g., ZmBz2 in maize, PhAN9 in petunia, AtTT19 in Arabidopsis, and VviGSTs in grape[37][73][74]. After heterologous overexpression of PhAN9 in Arabidopsis tt19 mutant, the anthocyanin pigment in seedlings was complemented, but no complementation was observed in the pale brown seeds of the tt19 mutant[37]. Moreover, PAs precursors were dispersed in the cytoplasm in tt12 mutant[71], but accumulated as membrane-like structures in endothelium cells of tt19, indicating that TT19 may function as a subsequent transporter to deliver the vesicles containing PAs precursors to the central vacuole[37][75]. VviGST3, a homolog to AtTT19, was highly expressed in the seed during grape berry development, and the complementation in seed color was observed in VviGST3 overexpressed tt19 mutant, suggesting that VviGST3 may be associated with the transport of PAs[76]. In addition, heterologous expression of pear PcGSTF12 in the Arabidopsis tt19 mutant showed that PcGSTF12 did not complement the mutant seed color to the normal brown, but it promoted the procyanidin A3 accumulation in the mutant and affected the transcription of PAs- and anthocyanin-related genes. The results of this study indicated that PcGSTF12 may be involved in the transport and accumulation of PAs and anthocyanins[77].
The vacuole is the final destination for the transportation of PAs, and the polymerization of PAs probably takes place here. Although little is known about the polymerization mechanisms of PAs, the enzymatic and nonenzymatic polymerization reactions are widely discussed[1][78][79]. Plant polyphenol oxidases, TT10 and plant peroxidases may take part in PAs polymerization and oxidation[1][64][80][81]. For instance, AtTT10 encodes a putative laccase that oxidizes epicatechin into yellow or brown oligomers, which may involve in PAs oxidative browning in the seed testa [64].
PAs have attracted attention not only because they possess beneficial pharmacological properties but also because of their special role in the regulation of fruit quality and plant defense. The principal focus of research in the biosynthetic pathway and regulatory mechanism of PAs has been explored for years, and some encouraging progress has been made. However, there are still many issues that need to be further explored. The external environment in which plants live is constantly changing, there is a diversity of species, and the complex genetic background of woody plants, results in different regulatory mechanisms of PAs in various species in response to developmental cues and various stresses (Figure 3). In all species analyzed to date, a large number of MYBs regulate the synthesis of PAs that are involved in plant response to stress and signals. Nevertheless, the bHLH and WD40 cofactors of the MBW complex are also involved in the regulation of other pathways, so their specificity for PAs-regulation is less than that of MYB TFs, and few studies have focused on bHLH TFs, WD40 protein, and the proteins from other families that also participate in the biosynthesis of PAs. The highly induced transcription and accumulation of PAs are considered to be a systemic response of plant to adverse conditions[82]. Further work needs to reveal how the biotic and abiotic stresses trigger the transcription of these TFs, how the transcriptional and post-transcriptional regulation modifies these TFs, and how the crosstalk between these TFs and various signals and hormones is conducted.
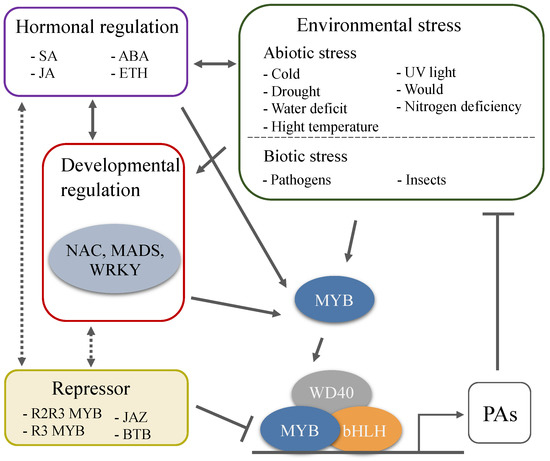
Figure 3. Environmental and developmental regulation of PAs biosynthesis through the MBW complexes. A large number of MYBs regulate the synthesis of PAs that are involved in plant response to stress and signals. Plant hormones are response to the environmental and developmental cues, and are the key factors involved in PAs biosynthesis. Environmental stresses affect the accumulation of PAs, and the high levels of PAs further enhance plant tolerance to extreme environmental challenges.
Metabolic engineering and molecular biology methods can be used to produce PA-rich crops that can enhance the tolerance of crops to various environmental stress, and improve the quality and nutritional value of crops. PAs have a great impact on the sensory properties of fruits by contributing to the astringency and bitterness, and may also influence the digestive system in humans and animals[1][83]. For example, the accumulation of PAs in persimmon fruit leads to astringency[84], which brings unpleasant tactile sensation. Understanding the regulation of PAs biosynthesis network can provide further insights into more targeted gene modifications, and are of great importance for breeding. However, it should be noted that one factor might affect the biosynthesis of many other metabolites. For example, the overexpression of VvMYBPA1 in tobacco upregulates PAs metabolism while downregulating anthocyanin biosynthesis[85]. Therefore, the comprehensive and in-depth analysis of the molecular regulation mechanism of plant resistance mediated by PAs has a positive significance for improving our knowledge of regulatory network controlling PAs synthesis and provides a theoretical foundation for genetic improvement and breeding of the plants with moderate levels of PAs.