Regulation of iron availability is an important part of cellular homeostasis. The bioavailability of iron is limited by the insolubility of ferric salts, while the excess of free iron leads to oxidative stress. Sequestration of iron ions within cells is mediated by ferritins, a widely distributed and conserved protein family found in all domains of life. Ferritins are organized in complexes consisting of 24 subunits forming a hollow sphere that is able to accumulate and store up to 4500 Fe
3+ atoms as ferrihydrite and, thereby, serve as a buffer to sequester excessive iron
[1]. When iron ions are needed, lysosomal degradation leads to their release from the ferritin complexes. Mammalian ferritin is composed of heavy (H) and light (L) chains. The H subunits are responsible for rapid detoxification of iron by ferroxidase activity
[2][3], and the L subunits facilitate iron nucleation, mineralization, and long-term storage
[4][5]. Members of the ferritin-like protein superfamily are involved in multiple cellular metabolic pathways, the redox-stress resistance, DNA replication, chlorophyll biosynthesis, endospore coat formation, fatty acid metabolism, tRNA modification, monooxygenase reactions, detoxification, and biomineralization
[6][7][8]. Ferritins are used as delivery vehicles for iron and drugs, as well as biomarkers of various diseases, so the main important functions of ferritin have been studied in vertebrates. Knockout of the mouse H-ferritin (MoHF) is embryonically lethal, and inactivation of MoHF makes the cells, especially myeloid, more sensitive to oxidative damage
[9][10][11]. Ferritin synthesis is stimulated during the development and cell differentiation, inflammation, and tumorigenesis. A decrease in H-ferritin can induce epithelial-to-mesenchymal transition of mammalian tumor cells through the TGF- β1 pathway
[12][13][14]. Hypoxia inducible factor A (HIFA) can directly bind with hypoxia response element (HRE) in the promoter region of human L-ferritin (HuLF) to enhance its expression thus regulating epithelial-to-mesenchymal transition of glioma
[15]. Cell death by ferroptosis is characterized by the iron-dependent accumulation of reactive oxidized lipid species and depends on the regulation of iron storage and ferritin expression
[16]. Sensitivity to ferritinophagy and ferroptosis varies between different cell types. The resistance to ferroptosis is provided by the prominin2-MVB-exosome-ferritin pathway and has consequences for iron homeostasis and cancer
[17]. The most specialized mammalian cells which are highly sensitive to iron deficiency and ferroptosis are the neural cells
[18]. They utilize excessive iron under certain conditions (magnetic resonance imaging) faster than other body cells
[19]. Invertebrate ferritins perform some unique functions that are not seen in the ferritins found in vertebrates. Ferritin of
Apis mellifera (Honeybees) participates in magnetite formation in their trophocytes
[20]. The ferritin-like superfamily proteins are associated with immune functions in the marine invertebrates
[21], and ferritin ChF of the marine tubeworm
Chaetopterus sp. is involved in the production of bioluminescence in the secreted mucus
[22]. The crystal structure of ferritin from the marine bivalve mollusk
Sinonovacula constricta predicted the iron-binding sites in the 3-fold channel, ferroxidase center, and putative nucleation sites, similar to the mammalian ferritins
[23]. Exploring the functions of ferritins in ancient animals is of special interest.
Sponges (phylum Porifera) are benthic animals, and likely represent the oldest phylum of the existing Metazoa. Sea sponges of the sub-tidal zone are well adapted to changes in the temperature and oxygen content and provide a unique model for studying the cell adaptation processes in animals
[24]. The sea sponges grow only in the presence of iron ions
[25]. Some bacterial symbionts use iron receptors or enzymatic systems to extract iron from sponges
[26][27]. These interconnections are genetically inherited and could be activated by low iron concentrations in the environment
[28]. The main component of the sponge body is an aquiferous system that undergoes numerous rearrangements in the course of the sponge life cycle, as well as due to constant changes in the environmental conditions
[29][30][31]. The sponge body is represented by several types of cells that can be in different redox and metabolic cycle phases. The gradients of oxygen, metabolites, and proteins stimulate constant movements and transformation of cells. The sponge cells have the capacity to transition between multiple cell types similar to the transdifferentiating and stem cells of mammals
[32][33][34]. A unique feature of sponge cells is the ability to reaggregate and form functional primmorphs from dissociated cells. The experimental model of the sponge cells reaggregation was first introduced by Wilson in 1907
[35] to study morphological transitions in sponges and lately was exploited in many laboratories
[35][36][37]. It has been demonstrated that
Halisarca dujardini (cl. Demospongia) has a high regenerative capacity that combines protective and regenerative mechanisms
[38][39]. By using biochemical methods and transcriptomic analyses, we have previously described iron metabolic and heme biosynthesis/transport pathways and their association with the reaggregation process in two cold-water sea sponges of cl. Demospongia,
Halisarca dujardini (lacking spicules) and
Halichondria panicea (having spicules)
[24][40].
2. Ferritin Genes
In order to recover ferritins from studied sponges, we combined data from our published de novo transcriptomes for Halisarca dujardini and Halichondria panicea (NCBI project numbers PRJNA594150 and PRJNA594151) with draft genomic data for H. dujardini. Three ferritin sequences were identified in the transcriptomic data of H. dujardini, HdF1a, HdF1b, and HdF2, and one ferritin sequence was identified for H. panicea, HpF1. Using novel genomic data for H. dujardini, we found that the HdF1b gene is located 3538 base pairs away from the HdF1a gene in the same scaffold. In comparison with HdF1a, the HdF1b gene features only two nucleotide substitutions in the coding region, one of which does not change the encoded amino acid, and another one leads to an A35G replacement (corresponding to position 38 by conventional human ferritin HuHF numbering). The coding part of the HdF2 gene sequence differs markedly from the HdF1a/b genes and is shorter by 6 nucleotides.
The genomic features of three H. dujardini ferritin genes were compared with those of marine (Amphimedon queenslandica, Sycon ciliatum, and Oscarella pearsei) and freshwater (Ephydatia muelleri and Lubomirskia baikalensis) sponges, which have published genomes. At least two ferritin genes were found in each sponge species. All ferritin genes of H. dujardini and S. ciliatum are intronless, while genes of other species have two or three exons.
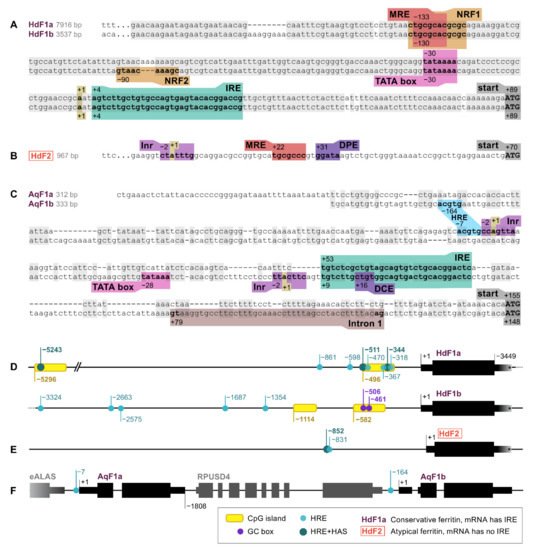
Ferritin genes of marine sponges H. dujardini, A. queenslandica, and freshwater sponge E. muelleri were screened for RNA and DNA regulatory sequences, such as promoter elements and motifs involved in the regulation of ferritin expression. The identified elements are shown in Figure 1. The 5′ upstream regions of HdF1a/b genes are highly similar up to 200 base pairs from the transcription start site (TSS) and, hence, contain a set of identified regulatory elements, whereas the pair of A. queenslandica AqF1a/b genes, which also originated from a recent duplication, have more diverse 5′ sequences (Figure 1A,C). The expression of HdF1a/b genes is controlled by a TATA-containing promoter located at position −30 upstream to the TSS. There is no TATA box in HdF2, instead, the CTATTT transcription initiation site (Inr) and downstream promoter element (DPE) were found at positions −2 and +31, respectively (Figure 1B). The promoter of AqF1a/b has a classical structure (Inr in AqF1a, Inr and TATA in AqF1b), and AqF1b features the downstream core element (DCE). EmF1 and EmF2 both have Inr. The H. dujardini 5′ upstream regions have CpG islands of length 204 and 206 bp (HdF1a), 206 and 269 bp (HdF1b), while short upstream regions of AqF1a/b and long upstream regions of EmF1 and EmF2 lack them. (Figure 1D,F). Two Sp1 transcription factor binding sites (GC box) were found in HdF1b at −506 and −461 inside a CpG island. There is a distal nuclear factor-κB (NFkB) motif located at position −5282 inside the CpG island of HdF1a. Metal-responsive elements (MRE) were found at various distances of the transcription start site, including proximal positions −133, −130, and +22 in HdF1a, HdF1b, and HdF2, respectively, while A. queenslandica ferritins lack them and E muelleri’s ferritins possess them only at distal positions (−6…−2 Kbp). Proximal MRE motifs found in HdF1a/b were contained within a larger motif of nuclear factor erythroid 2 (NFE2) -related factor 1 (NRF1), which is also present in EmF1 at −34. Interestingly, an NRF2 motif is identified in HdF1b, whereas in HdF1a it is disrupted by insertion of 3 bp. Hypoxia response element (HRE) motifs were found in all analyzed upstream regions, while more robust combinations of HRE and Hypoxia ancillary sequence (HAS), usually located by 7–15 bp downstream of HRE) were identified only in HdF1a (i.a. two are inside CpG island), HdF2 and EmF2 (Figure 1D–F). Weak CNC–sMaf binding element (CsMBE) motifs (also known as ARE and EpRE) were found at proximal positions in HdF1b, EmF2 and distal positions in HdF1a and EmF2. Other putative motifs including Myb box, ABRE, NICE and Sph1 box were identified. Iron-responsive elements (IREs), mRNA hairpin motifs which are bound by the iron-regulatory protein IRP1, were found in the 5′ untranslated regions of HdF1a/b, AqF1a/b, and EmF1, as well as in H. panicea HpF1 mRNA (Figure 1A,C).
Figure 1. RNA and DNA regulatory elements in the ferritin genes of H. dujardini (HdF1a, HdF1b, and HdF2) and A. queenslandica (AqF1a and AqF1b). (A–C) 5′ sequences of sponge ferritin genes. (D–F) Arrangement of sponge ferritin gene copies, CpG islands, Hypoxia response elements (HRE) and Hypoxia ancillary sequences (HAS). 3′UTR lengths of H. dujardini ferritins are only shown schematically since unlike the 5′UTRs they have not been experimentally confirmed; the assembled transcripts are longer than depicted in the figure. MRE, metal regulatory element; IRE, iron-responsive element; Inr, initiator of transcription; DPE, downstream promotor element; DCE, downstream core element.
The obtained transcriptome of H. panicea was supplemented with the data from another studies (NCBI project PRJNA394213, sample SAMN07484311; ENA project PRJEB43257). The only form of ferritin, designated earlier as HpF1 (NCBI ID: QIZ30882.1), was found in all paired-end transcriptomic libraries. No other ferritin forms were found in the libraries except for contaminants. H. panicea ferritin transcripts form a large cluster of slightly different sequences . To estimate the degree of ferritin polymorphism, we selected a single reference transcript and analyzed “transcriptomic SNP” frequencies in its coding part, mapping the reads from each library. It turned out that transcripts from different libraries had 30–40 polymorphic sites with frequencies more than 5% in the coding region of 513 nucleotides, with most of the SNP sites being consistent between all libraries. Thus, H. panicea ferritins are highly polymorphic having non-allelic differences.
3. Ferritin Proteins
The H. dujardini ferritins HdF1a and HdF1b each have 169 amino acids and differ slightly in the predicted molecular weights (MW): 19367.59 Da and 19353.56 Da, respectively. The isoelectric point pI for both is 4.91. The sequence of HdF2 protein differs significantly from the HdF1a/b, showing only 49% identity (69% similarity) with them and is shorter by two amino acids. Its predicted parameters are MW of 19425.84 Da and pI 5.87. The H. panicea ferritin HpF1 is closer to HdF1a/b than to HdF2: 60% identity (76% similarity) for HdF1a/b and 39% identity (60% similarity) for HdF2. HpF1 has 170 amino acids and a predicted MW of 19561.03 Da and pI 4.95.
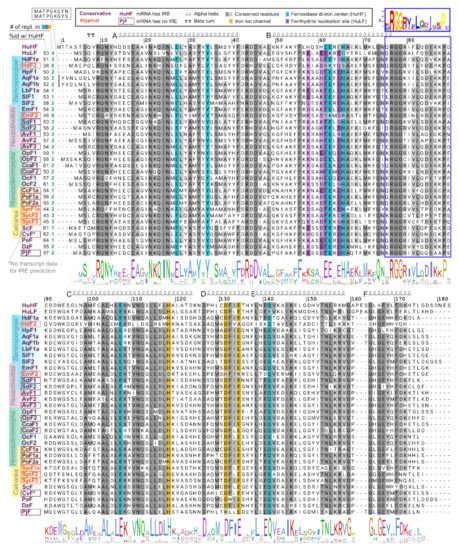
To compare functional domains of sponge ferritins, we constructed multiple amino acid sequence alignment for a set of sponge, human, and several marine invertebrate ferritins, which 3D structures have been recently published (Figure 2). H. dujardini and H. panicea ferritins show very high sequence similarity with other sponges and invertebrates and contain known conserved functional domains of H chain ferritins, namely the iron ion channel and ferroxidase di-iron center. The residues corresponding to ferrihydrite nucleation center, a domain usually attributed to L chain ferritins, do not follow the domain consensus in the majority of analyzed sponges. H. djuardini ferritin HdF2 is the least conserved and has substitutions in the key residues of functional domains: E27Q, E61S, E62K, H65S (ferroxidase di-iron center/ion binding site), and E134A (iron ion channel); all numbers follow conventional human HuHF numbering starting after initial methionine (Figure 2). The N-glycosylation (GlcNAc) site Nx[ST] (residues 111–113) was found in AqF1a, AvF1, AvF2, OpF1, OcF1, OcF2, CcF1a, PoF1a, PoF2a, and SycF3. The conserved protein kinase C phosphorylation site [ST]x[RK] (residues 144–146) present in freshwater sponge ferritins EmF1, SlF1a/b, LbF1a/b, and among other sponges: AvF3, OpF1, OcF1. All sponge ferritins lack the signal peptide for classical ER-Golgi secretion but have a non-classical endosome secretion pathway motif of 15 amino acids starting from xRGG in the BC interhelical region (framed with blue in Figure 2). A short turn between helices C and D (D126) is highly conserved in most studied invertebrates (Figure 2).
Figure 2. Domains and secondary structure of sponge ferritins. Alignment of ferritin amino acid sequences of H. sapiens, sponges of four classes, and five invertebrate species whose crystallographic data was recently obtained. Amino acids numbering corresponds to conventional human HuHF numbering starting after the initial methionine. Highlighted are residues of three ferritin domains: iron ion channel (yellow), ferroxidase di-iron center (cyan), and ferrihydrite nucleation site (purple). To the left of the alignment are shown identity percentage level with human HuHF and the number of replacements in three domains. HuHF secondary structure is shown schematically above the alignment (PDB ID: 3AJO). A motif for the non-classical endosome secretion pathway is framed with blue.
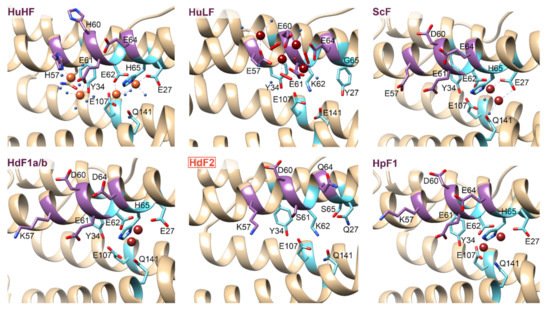
The 3D structures of H. dujardini, H. panicea, A. queenslandica, and E. muelleri ferritin domains complexed with iron ions were reconstructed by homology modeling using a structure of Sinonovacula constricta ferritin ScF (PDB ID: 6LP5) as a template, since it was the closest homologue with the known structure complexed with iron ions, having the sequence identity of 63.2%, 46.3%, and 55.1% with HdF1a/b, HdF2, and HpF1, respectively (Figure 3). Other possible templates included ferritins of marine worms Phascolosoma esculenta and Dendrorhynchus zhejiangensis (PDB IDs: 6LPD, 7EMK). The predicted models have good estimated quality (QMEAN scoring function values are 0.14, −1.77, and 0.08 for HdF1a/b, HdF2, and HpF1, respectively). In the HdF1a/b, HpF1, AqF1a/b, and EmF1 ferritins, the iron atoms in the structures are predicted to be connected only at the ferroxidase center. In HdF2 and EmF2, the iron-binding activity is not predicted at all. Transient heme binding was predicted for HdF1a/b, HdF2, HpF1, EmF1, and EmF2 ferritins for histidine residue at the position corresponding to L165 in HuHF.
Figure 3. 3D structures of the regions corresponding to ferroxidase di-iron center (cyan) and nucleation site (purple) of
H. sapiens ferritins HuHF and HuLF (PDB IDs: 4OYN, 5LG8), bivalve
Sinonovacula constricta ScF (PDB ID: 6LP5), and sponges
H. dujardini and
H. panicea ferritins. Sponge ferritins’ structures are modeled with SWISS-MODEL server
[41] using ScF as a template. Molecular graphics was prepared using UCSF Chimera software
[42]. Orange and brown spheres represent Fe
3+ and Fe
2+ ions, respectively. Amino acids numbers correspond to conventional human HuHF numbering starting after the initial methionine.
4. Phylogenetic Analysis of Sponge Ferritins
In order to reveal the presence of atypical non-conservative ferritins similar to HdF2 in other invertebrate species, we compiled a large dataset from annotated and unannotated transcriptomic databases, collecting 533 ferritin sequences belonging to 264 invertebrate species from more than 30 classes. Surprisingly, the hits found in all available Ctenophore databases represent only distant homologues of ferritin, therefore, no ctenophore sequences were included in the dataset. Among analyzed ferritins, 70% of insects’ and only 25% of other invertebrates’ sequences have signal peptides (SP), while all analyzed sponge ferritins lack them.
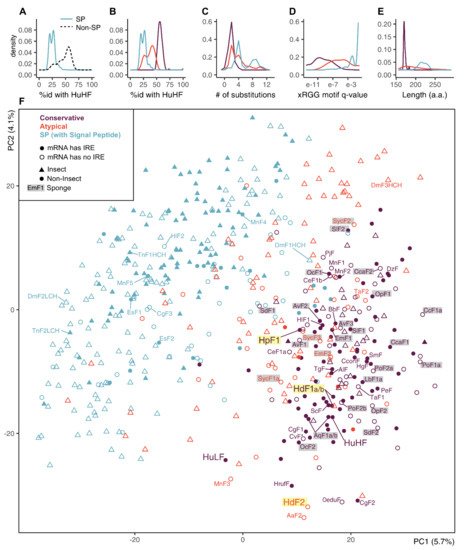
The SP-ferritins have low identity to human HuHF (Figure 4A). The SP-less ferritins generally have higher identity to HuHF, but the distribution of identity levels is skewed, suggesting non-uniform conservation and the presence of infraclasses. We defined two such infraclasses of SP-less ferritins, conservative and atypical (the atypical is defined as having less than 49% sequence identity with HuHF or more than five amino acid replacements in the three functional domains) (Figure 4B,C). The atypical ferritins occupy an intermediate position between the conserved non-SP ferritins and SP-ferritins in terms of the identity levels with HuHF, the xRGG motif enrichment, and the sequence length (Figure 4B–E). The proportion of conservative and atypical classes in analyzed ferritins differs for insects (8% and 23%) and other invertebrates (57% and 18%). In sponges, atypical ferritins are represented by HdF2 (37.7% identity with HuHF and 8 replacements), E. muelleri EmF2 (46.4% id; 9 repl.), and all S. ciliatum ferritins: SycF1a/b, SycF2, SycF3 (44.3–45.4% id; 2–3 repl.). Nearly all atypical ferritins lack iron-responsive element (IRE), the few exceptions can be considered as marginal cases of conservative ferritins (47–49% id; 2–3 repl.). The overall level of IRE presence is 47% for non-insect and 29% for insect ferritins. IREs are mostly found in the ferritin classes with the largest representation in the group: 63% of conservative ferritins outside of insects, and 41% of insect SP-ferritins .
Figure 4. Principal component analysis (PCA) plot on a set of animal ferritins built using 474 sequence features selected by unsupervised algorithms. (A,B) Distribution of identity percentages with HuHF in ferritins of two classes of SP and SP-less ferritins and after dividing SP-less ferritins into conservative and atypical classes. (C–E) Distributions of the number of replacements in the three functional domains, the xRGG motif q-value, and protein sequence lengths in the ferritins of three classes. (F) PCA plot of 533 analyzed invertebrate and human ferritin sequences built by selected feature subset and showing prior class labels. Sponge ferritins are highlighted with grey, H. dujardini and H. panicea ferritins are highlighted with yellow.
To identify possible common characteristics shared by atypical invertebrate ferritins, we performed dimensionality reduction on analyzed sequences’ features. First, a comprehensive protein sequence feature set was obtained that includes simple, grouped, and pseudo amino acid composition, composition/distribution/transition, Geary, Moran, and normalized Moreau-Broto autocorrelation features for each sequence. Next, this set was reduced from 6567 to 474 features (7.2%) combining top-100 features selected by five unsupervised feature selection algorithms. We did not infer new clusters but transferred our class labels to a PCA projection built for the selected feature subset. The PCA demonstrates a relatively clear differentiation between the SP-less and SP-ferritins, but the differentiation between conservative and atypical ferritins is indistinct (Figure 4F). The sponge ferritins, atypical HdF2, SycF1a/b, SycF2, conservative HpF1, SdF1, SdF2, SlF2 lie on the periphery of the conservative ferritin cluster, as well as the human HuLF. At the same time, atypical EmF2 and SycF3 are closer to the center of the cluster, together with the conservative HdF1a/b, AqF1a/b, and human HuHF. Bivalve ferritin ScF, which was used as a template for 3D modeling, is located very close to HdF1a/b.
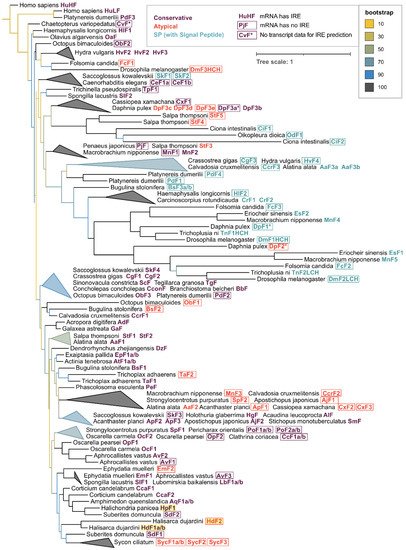
For phylogenetic analysis, the initial collection of 533 ferritins was reduced to 131 sequences while maintaining the representatives of all main classes since the majority of annotated ferritin sequences belonged to a single class Insecta (58% of the whole dataset). The phylogenetic tree built with the selected subset of 131 invertebrate ferritins with human ferritins used as an outgroup shows that the sequences tend to group not only by the respective taxonomy but also frequently with regard to the defined ferritin classes (Figure 5). Interesting examples of grouping by ferritin class are mollusks’ (including bivalve ScF) and Cnidarian conservative ferritins, atypical ferritins from different phyla of marine invertebrates and a large cluster of SP-ferritins. Sponge ferritins form a distinct cluster in the tree, with the freshwater species clustering together. Among the most evolutionary distant sponge ferritins are atypical HdF2, EmF2, ferritins of Calcarea sponge S. ciliatum; conservative HpF1, SdF2, and Hexactinellida sponge A. vastus AvF1, AvF2.
Figure 5. Phylogenetic tree of invertebrate and human ferritins. 131 representative ferritins from 56 species of 32 invertebrate classes along with human ferritins were aligned using clustalo algorithm
[43], the alignment was processed with Gappyout algorithm using trimal v. 1.2 software
[44] to remove variable ends and regions of signal peptide (132 core residues left). The tree was constructed using the Maximum likelihood approach with 1000 fast bootstrap resamplings with IQ-Tree v. 1.6.12
[45] and visualized using iTOL server
[46], with ferritin sequences of
Homo sapiens used as an outgroup. Triangles represent collapsed clades with upper and lower points showing the range of branch lengths inside a clade.
2.4. Ferritin Complexes
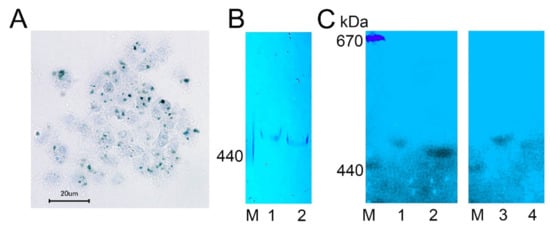
We visualized complexes containing ferric ions in H. dujardini cells by using specific Prussian blue staining (Figure 6A). The ferric complexes were observed in cells as optically dense areas of different sizes. Fractionation of the cell extracts by a native electrophoresis in polyacrylamide gels followed by staining with Prussian blue revealed ferritin complexes in both sponges, H. dujardini (lacking spicules) and H. panicea (having spicules) (Figure 6B). The H. dujardini ferritins migrated a little faster than that of H. panicea, but both complexes migrated slightly slower than the horse ferritin marker (440 kDa). As expected, the ferritin complexes in gels showed absorption of UV light near 360 nm wavelength (Figure 6C).
Figure 6. Ferritin complexes in sponges H. dujardini and H. panicea. (A) The ferric complexes detected in H. dujardini cells by Prussian blue staining. (B,C) Sponge cell extracts fractionated by native electrophoresis in polyacrylamide gel: staining with Prussian blue (B) and the UV 360 nm absorbance (C). Specimens of H. panicea collected in Autumn (1) and Summer (3). Specimens of H. dujardini collected in Autumn (2) and Summer (4). M, horse ferritin (440 kDa) and thyroglobulin (670 kDa).
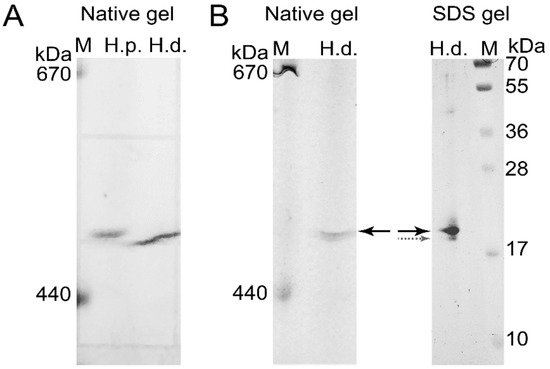
To confirm the presence of H. dujardini and H. panicea ferritins in the high molecular mass complexes detected in native gels, the corresponding bands were cut from the native gel stained with Coomassie blue without fixation (Figure 7A) and then analyzed by MALDI/TOF mass spectrometry. The HpF1, HdF1a/b, and HdF2 ferritins were identified in these bands. In addition, the ferritin band of H. dujardini (specimens collected in Summer) was cut from the native gel (Figure 7B), denatured in SDS-containing buffer and analyzed by the SDS-12% PAGE electrophoresis followed by Coomassie staining. One major band of approximately 20 kDa was revealed (Figure 7B) that well corresponds to a predicted molecular mass of HdF1a/b equal to 19.4 kDa. The MALDI/TOF mass spectrometry confirmed the presence of HdF1a/b in this band. The minor band of approximately 19 kDa that resolved by this analysis (Figure 7B) corresponds to H. dujardini neuroglobin (QEH04777.1).
Figure 7. Isolation of H. dujardini and H. panicea ferritins by electrophoresis in a native polyacrylamide gel for mass spectrometry. (A) The native polyacrylamide gel stained with Coomassie blue without fixation. (B) The band corresponding to H. dujardini ferritin complex (marked by solid black arrow) was taken from the native gel, denatured, subjected to SDS-12% PAGE electrophoresis, and then stained with Coomassie blue. The major and minor bands indicated by solid black and dotted grey arrows were taken for MALDI/TOF mass spectrometry. M, horse ferritin (440 kDa) and thyroglobulin (670 kDa). H.p., H. panicea; H.d., H. dujardini.
2.5. Subcellular Localization of Ferritins
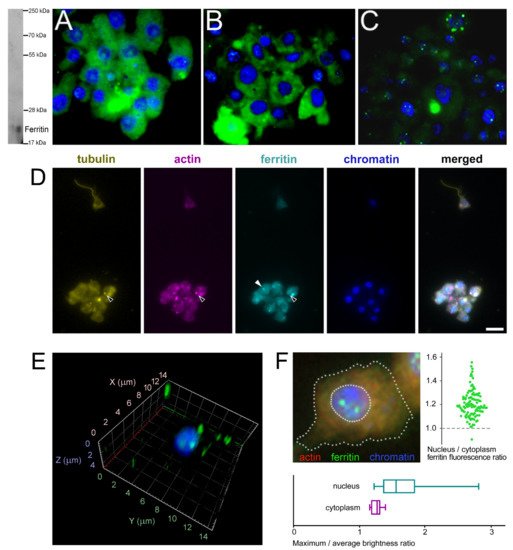
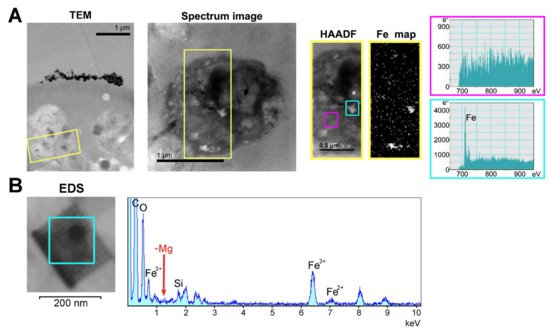
The subcellular localization of H. dujardini ferritin was studied using an immune fluorescence assay. The cytoskeletal proteins, actin, and tubulin, were visualized by specific antibodies. Ferritin shows predominantly diffuse distribution in the nucleus and cytoplasm of sponge cells (Figure 8A,B) with a number of speckles producing bright fluorescent spots under immunostaining (Figure 8C). The ratio of ferritin fluorescence in the nucleus and cytoplasm was 1.21 ± 0.11 (0.01) {mean ± Sd (Se)} for 114 cells (Figure 8F, swarmplot). This value indicates roughly similar spreading of ferritin between nucleus and cytoplasm in sponge cells where the nuclear area does not differ significantly in size from the cytoplasmic area in most cells. However, ferritin is distributed more heterogeneously inside the nucleus than in cytoplasm as illustrated by the higher ratio of maximum brightness to average brightness in the nucleus than in cytoplasm (nucleus: 1.80 ± 0.64 (Sd) vs. cytoplasm: 1.30 ± 0.09 (Sd) (Figure 8F, boxplot)). Moreover, the standard deviation values clearly indicate the higher variation in the nuclear staining than in cytoplasmic staining. The colocalization of ferritin with actin and tubulin was observed in cells with or without flagella as well as in dissociated and reaggregated cells (Figure 8D). The bright spots of ferritin complexes varied in numbers from 1–2 to 8 or more and were located largely in the nucleus (Figure 8E,F). The nuclear speckles of ferritin were more pronounced in cells at the beginning of sponge growth (specimens collected in August) (Figure 8C). The presence of the storage iron form, oxidized ferric ions (Fe3+), in the electron-dense subcellular structures presumably associated with ferritins in sponge cells was directly confirmed by the transmission electron microscopy accompanied by spectral analysis (Figure 9). The Fe3+ iron signal was clearly recognized in the spectrum of the electron-dense area lacking the magnesium atoms. This result confirmed the accumulation of ferric ions in specific subcellular structures in sponge cells revealed by staining with Prussian blue (Figure 6A).
Figure 8. Subcellular localization of ferritin in H. dujardini cells. (A–C) Ferritin immune staining. Ferritin (green), Chromatin (blue). Western blot on the right shows the antibody specificity. Staining of cells collected at the end of sponge growth in Autumn (A), and at the beginning of sponge growth in Summer (B,C). The focal planes of maximum intensity presented. (D) Sponge cells, quadruple stained for tubulin, actin, ferritin and chromatin. Black arrows, cytoplasmic ferritin colocalized with both tubulin and actin; white arrow, cytoplasmic granule of ferritin without visible signs of colocalization with cytoskeletal proteins. Scale bar 10 μm. Confocal microscopy of intranuclear ferritin in the isolated nuclei, 3D reconstruction. (E) Ferritin (green), Chromatin (blue). Intranuclear ferritin granules are co-purified together with nuclei. (F) Sponge cell, triple stained for actin, chromatin and ferritin (dashed lines denote boundaries of the cell and the nucleus). Swarmplot shows the ratio of average fluorescence levels between ferritin localized in the nucleus and in the cytoplasm of each cell, N = 114. Boxplot compares the ratio of the maximum to the average fluorescence level of ferritin localized in the nucleus and in the cytoplasm of each cell, N = 114.
Figure 9. Identification of the ferric ion-containing granules in
H. dujardini cells. (
A) Transmission electron microscopy of the fragment of sponge cell with inclusions. HAADF and Fe map panels represent the area outlined in yellow on the panel Spectrum image which in turn is a section of the TEM panel. The spectra outlined in cyan and magenta were taken from the corresponding outlined areas in the HAADF panel. Scale 1 μm. (
B) EDS-analysis of the region where the presence of iron atoms was confirmed by the HAADF method. TEM image of this area in higher resolution is shown in
Figure S7.
2.6. Expression of Ferritins and the Ferritin Associated Factors during Different Periods of the Annual Cycle
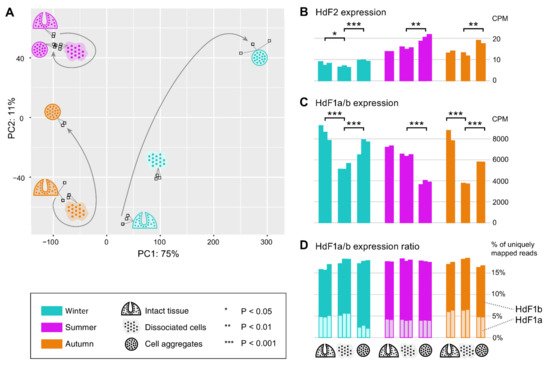
In order to reveal possible involvement of ferritin and other iron metabolic proteins in the morphogenetic processes in sponges, we carried out transcriptomic sequencing for samples of H. dujardini collected at different periods of their life cycle: Winter (the beginning of spermatogenesis and oogenesis), Summer (the beginning of sponge body tissue growth), and Autumn (the end of sponge body tissue growth), and at different reaggregation states (intact sponge body tissues, dissociated cells, and reaggregated cells). Our previous data for Autumn samples (NCBI PRJNA594150, samples SAMN13506244‒SAMN13506251) were combined with new data for the Summer and Winter samples that were deposited to the National Center for Biotechnology Information (NCBI PRJNA594150, samples SAMN20337311‒SAMN20337327). The average number of single-end 50 bp reads per sample was 28 M with the total number of 476 M reads for 17 new samples of H. dujardini. On average, 99.8% of reads passed the quality filter, of which 95.1% were successfully aligned to the transcriptome assembly constructed earlier (NCBI TSA: GIFI00000000.1). Although the studied growth periods are represented predominantly by somatic, non-reproductive cells in the sponge body, the biological coefficient of expression variation showed the value of 0.73, thus confirming the transcriptional diversity of the studied samples. The PCA plot reveals clusters of samples associated with the aggregation states of sponge cells from each of the life cycle stages (Figure 10A). Additionally, the samples associated with the beginning of sponge growth (Summer) cluster more tightly than the other life cycle stages. The reaggregated cells from the Winter and Autumn periods tend to separate from the other aggregation states of the corresponding seasons.
Figure 10. Expression of ferritin genes during the dissociation/reaggregation processes in sponge H. dujardini during the different periods of the annual cycle. (A) Principal component analysis (PCA) plot of RNA-Seq samples colored by the life cycle stages (Winter, cyan; Summer, purple; Autumn, orange) and marked by the pictograms of the stages of the dissociation/reaggregation experiment. (B,C) Ferritin HdF2 and HdF1a/b transcript expression in CPM (calculated by edgeR after normalization), each bar represents one replicate. Statistical significance is shown for BH-adjusted p-values of the t-test carried out for replicates of each period. Since the HdF1a and HdF1b copies are nearly identical, their expression is shown in total (reads were mapped to one transcript). (D) Stacked barplot showing the percentage of the reads mapped uniquely to HdF1a (light boxes) or HdF1b (dark boxes) gene copy, out of the total number of reads mapped to these copies, that could be used as an estimate for the transcriptional ratio between two copies.
Ferritins HdF1a/b are among the most expressed genes in H. dujardini, along with such genes as actin and tubulin. In the cells of intact sponges, their expression was similar in all studied life periods (Figure 10C). The HdF1a/b expression markedly decreases in the dissociated cells of sponges collected in Winter and Autumn, and partially recovers during reaggregation. In contrast, the HdF1a/b expression does not change significantly after dissociation in samples collected in the Summer season but markedly decreases under reaggregation (Figure 10C). Interestingly, the relative proportion of a/b copies was similar in all studied samples, except for the aggregated cells of the Winter samples, where the proportion of HdF1a was markedly decreased (Figure 10D). The HdF2 expression in the Winter samples, associated with the onset of spermatogenesis and oogenesis, was markedly lower than in the Summer and Autumn samples. The aggregation of cells was accompanied by an increase in HdF2 expression in all samples (Figure 10B).
The expression levels of other factors involved in iron metabolism, heme biosynthesis and transport, response to hypoxia and ferroptosis were lower than that of ferritins HdF1a/b. Actual expression levels are dependent on the seasons when the sponge samples were collected and affected by the dissociation/reaggregation processes. Most factors involved in the iron and heme metabolism (SQSTM1, NAALAD2-like 1, IPR1/ACO1, NFKB1, GAPDH, ABCG2, HRG1-like, globin NGB) have a higher expression level in body tissues in the Summer period than in Winter or Autumn. The same was true for the hypoxia factors HIFa/SIM-like 2/3, anti-apoptotic protein BCL2, and ferroptosis factors, prominin 1, TGFBR1-like 2 and glutathione peroxidase GPX-like 2. Expression of BCL2 increases under reaggregation of cells, meanwhile SQSTM1 expression increases during reaggregation only in Winter and Autumn samples but remains unchanged in Summer samples. The metal-regulatory transcription factor 1 (MTF1) linked to the regulation of ferritins is expressed in sponge bodies at a low level at all seasons, but it is markedly induced during cell reaggregation in Winter samples. Reaggregation also increases expression of the heme transporter ABCB6 in all seasons and that of transporter ABCG2 in Winter. Among the heme biosynthesis enzymes (ALAS, ALAD, and FECH/hemH), expression of ALAS and ALAD decreases during reaggregation in all samples, but increases for FECH/hemH in Winter samples. Expression of neuroglobin NGB changes differently during dissociation/reaggregation processes in sponges collected in different seasons. Reaggregation is accompanied by an increase in NGB expression in the Winter and Autumn samples but not in the Summer samples. Another expression pattern was observed for Cathepsin D and Casp3: the basal expression level in sponge tissues in Winter and Autumn was lower than that in Summer, and it markedly decreases during the reaggregation process. The differential expression pattern of factors involved in iron metabolism at different periods of sponge life cycle briefly described above reveals intricate regulation of morphogenetic processes in sea sponges.
2.7. Ferritin Superfamily Members of the Microbial Community of H. dujardini
Bacterial representatives of the ferritin superfamily were identified in the transcriptomic data of H. dujardini, namely, rubrerythrins, non-heme ferritins, bacterioferritins, and DNA starvation/stationary phase protection proteins (Dps). The top hits obtained via blastp by querying against bacterial ferritin sequences of NCBI Protein database have good sequence coverage (68–100%) and identity values (44.6–100%). All these microbial symbionts were Gram-negative bacteria that belong primarily to Alphaproteobacteria and Gammaproteobacteria in the phylum Proteobacteria. Some of them could be identified at the family level (Rhodospirillaceae, Rhodobacteraceae, Rhizobiaceae, Xanthomonadaceae, Thiotrichaceae) or at the genus and species levels: Magnetospira sp., Magnetospirillum gryphiswaldense, Desulfovibrio desulfuricans. Labrenzia sp., Halovulum dunhuangense, Bosea sp., Pseudomonas asplenii, Albidovulum inexpectatum, Ruegeria pomeroyi, Brevundimonas bullata, Kiloniella litopenaeus, Nisaea denitrificans, Oceanibaculum nanhaiense, Chryseolinea flava, Notoacmeibacter marinus, Endozoicomonas sp., Vibrio cyclitrophicus, Magnetovibrio sp. Interestingly, the expression of ferritin superfamily members of many bacterial phyla, except for Cyanobacteria, showed lower levels in the sponge bodies collected in Summer compared to sponges collected in Autumn.