Doubly uniparental inheritance (DUI) of mitochondrial DNA (mtDNA) in bivalve mollusks is one of the most notable departures from the paradigm of strict maternal inheritance of mtDNA among metazoans.
1. Introduction
Over 100 species of bivalved mollusks are known to exhibit the unusual system of doubly uniparental inheritance (DUI) of mitochondrial DNA (mtDNA)
[1]. DUI is an inheritance pattern characterized by two sex-associated lineages by which mtDNA is transmitted, a male lineage and a female lineage
[2]. Typically, the male-transmitted mtDNA type (or M-type) is the exclusive type present in sperm whereas the female-transmitted type (or F-type) is the exclusive (or at least vastly predominant) type present in eggs and in somatic tissues of females
[3]. The F-type is also the more abundant type in male somatic tissues, whereas the M-type is typically present in lower amounts
[4]. DUI was first described in marine mussels (genus
Mytilus) approximately 30 years ago
[5][6]. In addition to marine mussels, DUI has been found in marine clams of the order Veneroida, freshwater mussels of the order Unionoida, and nutshells of the order Nuculanoida; however, the phylogenetic position of Nuculanoida (e.g.,
Yoldia) within or outside of the Protobranchia is controversial
[1][7]. In a recent review of current research in the area of DUI, Stewart and colleagues
[8] discussed the following topics: whether DUI evolved once or multiple origins, the challenges of annotating mtDNA genomes of DUI species, whether there is a link between the male- and female-transmitted mtDNA genomes and sex determination, what possible role(s) non-oxidative phosphorylation mitochondrial genes (i.e., open reading frames or ORFs) may play in freshwater mussels, and whether or not there are obvious evolutionary advantages of DUI, among other questions. One of the other points of discussion briefly considered was the possible function of conserved sequence motifs and sperm transmission elements (STEs) in the mitochondrial genomes of marine mussels. In this paper, we focus on this point by extending the analysis of Robicheau et al.
[9] to search for conserved DNA sequence motifs that could represent possible sperm transmission elements as first described in Kyriakou et al.
[10].
Within mitochondrial genomes, the search for molecular signals that may play a role in their sex-associated mode of transmission is facilitated by a particularly curious phenomenon best studied in marine mussels. As directly documented in several species within the blue mussel genus
Mytilus, and inferred in several other members of the Mytilidae through genome and phylogenetic analyses, the male- and female-transmitted genomes in DUI positive taxa occasionally recombine (reviewed in
[11]). While all manner of combinations of recombinant genomes are likely possibly, several known examples consist of recombinants composed of a mostly F-type genome (including the F-type protein coding genes,
12S and
16S ribosomal RNA genes, and most or all F-type transfer RNA genes) and an inserted control region from an M-type genome (e.g.,
[12]). Although there is variation in terms of exactly which additional adjacent portions of the male genome may also be recombined (e.g.,
[13]), this type of recombination event can result in a mostly female-type mitochondrial genome that is transmitted in a father-son pathway via sperm
[14]. Such recombinant genomes have been referred to as recently masculinized (RM) genomes and the event is described as a “role-reversal,” which refers to the switch in sex-associated transmission for the majority of genes in these recombinant mitochondrial genomes
[11]. An obvious conclusion to be drawn from the study of these recombinant genomes is that there could be a molecular signature (for e.g., a DNA motif) present in the M-type control region that facilitates its transmission from male parent to male offspring.
As summarized by Kyriakou et al.
[10], there are three possible mechanisms that could result in the M-type dominating transmission in sperm while the F-type remains the predominant type found in somatic tissues of males: (i) only mitochondria from the male parent enter the primordial germ cells (PCGs) that will go on to populate the spermatogonia; (ii) mitochondria from both parents enter the PCGs but the M-type mitochondria experience more rapid replication and, in effect, outcompete the F-type mitochondria in this arena; and (iii) the F-type mitochondria are actively eliminated from the PCGs leaving only the M-type, which must also have entered the relevant blastomere in early development. Using electrophoretic mobility shift assays of gonad extracts from Mediterranean sea mussels (
Mytilus galloprovincialis), Kyriakou et al.
[10] provided evidence that there is a DNA sequence motif in the control region of the M-type that forms a DNA-protein complex. These authors also demonstrated that this DNA-protein interaction occurs in the perinuclear space within male gonad cells, and they proposed that these perinuclear mitochondria are protected from general degradation of cytosolic mitochondria that results in the destruction of the F-type mitochondria and their mtDNA genomes. Kyriakou and colleagues thus argued that the third scenario above, i.e., the destruction of the F-type and the preferential survival of the M-type, is the most likely of the three possible mechanisms underpinning the DUI system, at least in marine mussels
[10].
Relatively little experimental or in silico work has been performed on mytilids over the past several years to follow up on the aforementioned work by Kyriakou et al.
[10]. Robicheau et al.
[9] used the STE signature of Kyriakou et al.
[10], a 22–23 bp conserved DNA sequence string, to search for putative STE motifs in the highly divergent male and female-transmitted mitochondrial genomes of the horse mussel,
Modiolus modiolus [15]. Overall, this work suggested that divergent, yet still recognizable, STEs may be found when less stringent search parameters are implemented during alignment procedures (e.g., by converting sequences to simply purines and pyrimidines). Indeed, a putative STE in the control region of the male mitochondrial genome of
M. modiolus was identified using this more relaxed search approach
[9]. Hence, the identification of homologous DNA sequence motifs that could be involved in the protection and (or) sequestration of the M-type mitochondrial genomes remains a feasible area for discovery despite the STEs’ rather short nucleotide signature.
In addition to the obvious relevance for understanding the evolution and maintenance of DUI in bivalve mollusks, studies of the molecular signatures involved in precisely controlling the nature and extent of mitochondrial heteroplasmy in animal cells have broad applicability in cell and molecular biology as noted by
[16]. Mitochondrial heteroplasmy is implicated in human diseases, some quite severe, affecting a number of organ systems
[17][18]. Teasing apart the mechanism(s) of the mitochondrial dynamics of the male- and female-transmitted mitochondrial genomes in bivalves could shed light on pathways of mitochondrial biogenesis (e.g., of the role of the nuclear-encoded gene PCG-1α
[19][20] and mitophagy (e.g., the PINK1/Parkin axis that ultimately activates mitophagy of select mitochondria in animals
[21]).
2. Mitochondrial Genomes of Bivalve Mollusks
The first extensive analysis of putative sperm transmission elements (STEs) in bivalve mollusks with the DUI system for mitochondrial DNA comes from the study by Kyriakou et al.
[10], which focused on
Mytilus spp. Accordingly, we used the 22 bp motif identified by those authors as a starting point to search for STEs in other bivalve mitochondrial genomes. As a critical first step, we needed to specify the search parameters to find a similarity threshold range that would minimize both false positive matches (i.e., searches that falsely identify non-homologous regions as matches due to low stringency) as well as minimizing false negative searches (i.e., searches with a stringency set so high that no or virtually no homologous regions were identified). As outlined earlier, a threshold of T = 56 for the known motif signature along with a sliding window length of 22 bp met these criteria (
Figure 1a,b).
Using the calibrated sliding window method above, we were able to identify a total of 23 STE-like signatures associated with LURs from the 157 mitochondrial genomes screened.
Figure 1c shows the nucleotides identified as having homology to the known STE motif (pink). The 30 bp upstream from each signature (with adenines highlighted yellow) is also shown in
Figure 1c. Out of the total LUR-associated putative STEs, four are previously known from Kyriakou et al.
[10] (Map Codes 3, 4, 5, and 8) and one is the known STE motif (★), these are highlighted grey (
Figure 1c). The STE signature from
M. trossulus M (Map Code 8) was manually added to the analysis, as its relatively low sequence similarity makes it interesting for comparative purposes, yet it could not be identified using our search strategy (hence, this motif represents a true positive that falls below the threshold of detection). The degree of sequence similarity between the putative STE motifs of
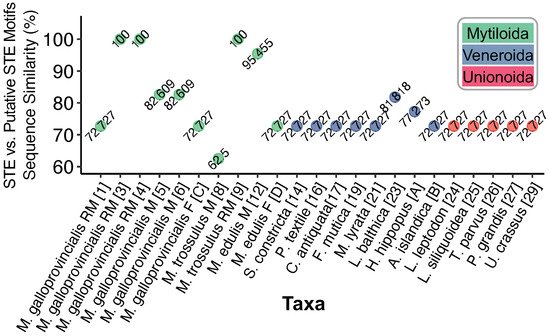
Figure 1c and the known STE is shown in
Figure 2. Putative motifs range in sequence similarity from 62.5% (15/24bp) to 100% or 22/22bp.
Figure 2. Degree of sequence similarity between STE-like signatures and the known 22 bp STE motif from Kyriakou et al.
[10]. Nucleotides were aligned using MUSCLE
[22].
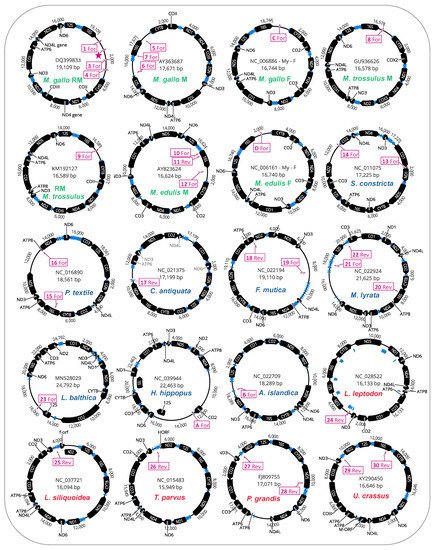
We further investigated whether the mitochondrial genomes housing putative STE motifs in their LURs also displayed evidence of additional STE homology to other parts of their mitochondrial genomes (i.e., outside the LURs). The full genome maps for protein-coding genes, tRNAs, and putative STE motif signatures are given (
Figure 3). As illustrated, for many of the genomes [12/20 mtDNAs] these motifs were only localized to the unassigned/control region (
Figure 3). Interestingly, for the eight genomes with additional motifs outside the LURs, these were in many cases on the complementary DNA strand to the protein-coding or rRNA gene they were identified in and were usually associated with either
16S (often), as well as
nd5,
atp6, or
co1 (less often;
Figure 3). The localization of many putative STE motifs to LURs is interesting given that nearly two decades ago, Burzyński et al.
[23], Zbawicka et al.
[24] and Cao et al.
[25] suggested that genetic signatures associated with DUI signaling were probably to be found in the non-coding portion of mtDNA as these unassigned regions or control regions, as they are variously called, are the mtDNA regions primarily associated with recombination events leading to the formation of masculinized mitochondrial genomes in the family Mytilidae.
Figure 3. Mitochondrial genome maps for bivalve species exhibiting putative STE motifs localized to large unassigned regions in mtDNA. STE-like signatures are identified by map codes within boxes (these correspond to codes in Figure 1c) and can either be found on the Reverse (Rev) or Forward (For) mtDNA orientation. M. gallo refers to M. galloprovincialis.
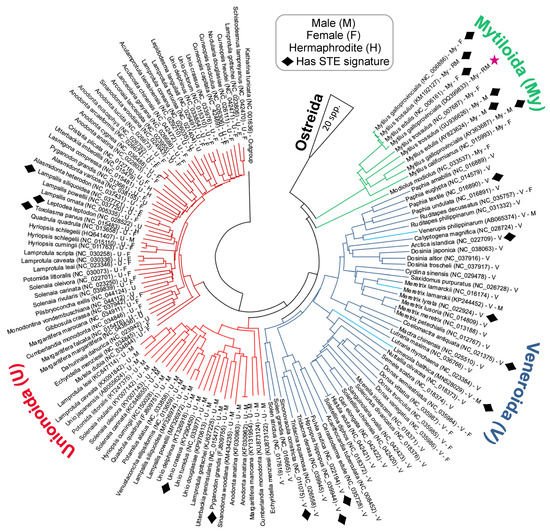
The phylogenetic distribution of bivalve mitotypes exhibiting putative STE signatures within their LURs is shown in the tree diagram presented in
Figure 4. It is important to note that the Neighbour-joining tree in
Figure 4 is based on the alignment of complete
co1 sequences. Although the tree is computationally quite simple, it nevertheless does an adequate job of representing the known broader evolutionary relationships previously indicated in the literature
[1][26]. Overall, the phylogeny indicates that several distantly related clades from across the Unionida, Veneroida and Mytiloida have signatures similar to the known 22 bp STE motif (
Figure 4). For those in Unionoida and Veneroida, signatures were found in thirteen species: three in DUI paternal genomes, two in DUI maternal genomes, one in an SMI hermaphroditic genome, and seven in genomes of unknown inheritance pattern for which we suppose are maternally-transmitted mtDNA (
Figure 1). Despite our widespread finding of this signature, we did not observe their presence among taxa as a monophyletic group, nor were the signatures limited to male-transmitted genomes (
Figure 4). For both trends, we must consider more closely that recombination between DUI mitotypes may obfuscate evolutionary patterns. As indicated earlier, recombination and masculinization of mtDNA genomes were demonstrated in the Mytilidae
[13][27][28], and recombination and role-reversal have been events implicated in the other DUI positive bivalve orders
[7][11]. Some of the male-transmitted mitochondrial genomes that were used during our analysis were known to be chimeric and, indeed, all M-types could be chimeric or derived from a chimeric ancestor. This means that the putative STE motifs are being mapped onto a genome tree rather than a gene tree. If we could confidently determine that recombination had occurred repeatedly and could reconstruct the recombination events throughout the hundreds of millions of years of bivalve evolutionary history, then we could determine whether the STE motif is monophyletic. Monophyly of the STE motifs across all bivalves would be strong evidence that the phenomenon evolved once. However, given the extremely long time periods involved, and the extremely small length of the putative STEs, reconstructing the evolutionary history of such events with greater confidence than we present here is highly unlikely. On a similar note, it is also possible that the putative STE motifs localized to F-type genomes could be the product of failed masculinization events (as suggested by the mt genomes analyzed in
[13][29]), or alternatively, they may instead represent non-functional STEs that have yet to acquire enough nucleotide differences via mutation to appear dissimilar to the functional STE (as implied by sequences in Kyriakou et al.
[10] for the
M. galloprovincialis F-type genome). Accordingly, the presence of STE-like signatures in F-types does not allow us to definitively argue that DUI arose only once, in an ancestor to the Mytiloida, Unionoida, and Veneroida orders. If the DNA sequence motif of the STE was only found in sperm transmitted genomes and could be shown to be present in the Mytiloida, Unionoida, and Veneroida, then the most reasonable explanation would be that the phenomenon had evolved only once. While this is still the most parsimonious explanation, it is possible that the M-type sequestration mechanism evolved independently three or more times in the history of the Bivalvia. Analysis of additional complete sequences of M and F mitochondrial genomes in the future should help test hypotheses of putative cases of recombination and masculinization within particular bivalve orders, which should, in turn, shed more light on the roles of STEs and uncharacterized mitochondrial open reading frames (ORFs) in the origin and maintenance of DUI across the Bivalvia
[8].
Figure 4. Taxonomic distribution of STE-like signatures discussed in this study. The phylogeny is a Neighbor-joining tree with 1000 bootstrap replicates reconstructed in MEGAX
[30][31]. Full-length
co1 genes were aligned using MUSCLE
[22] and trimmed to a conserved start/end nucleotide. ★ = Known STE motif from Kyriakou et al.
[10]. The
Hyriopsis schlegelii male sequence is likely an F-type genome derived from a male individual
[32].
Despite these aforementioned challenges in understanding the evolutionary history of STEs, our search for putative STE motifs amongst a larger set of bivalve mitochondrial genomes has allowed for two major steps of progress in our understanding: (i) our analyses suggest that the STE signature may represent a unifying feature that is present within LURs from all three of the major lineages of bivalves exhibiting DUI (clams, freshwater mussels, and marine mussels) despite its origins still remaining unclear; and (ii) using the putative STEs identified herein, we are now positioned to design targeted electrophoretic mobility shift assays to detect DNA-protein complexes in Unionoida and Veneroida as was done for Mytiloida
[10]. The most promising lineage currently appears to be
Limecola balthica given its 81% similarity to the 22 bp STE motif (
Figure 2). Moving forward, if the same DNA-protein complexes, and localization of perinuclear mitochondria can be demonstrated in the Unionoida as well as the Veneroida, this would be further strong evidence that the DUI phenomenon evolved once, but that there were subsequent role-reversal/recombination events in the ancestors to these two orders of bivalves as well.