
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | A.S.N.. Reddy | + 8314 word(s) | 8314 | 2021-04-25 10:53:38 | | | |
| 2 | Camila Xu | -2 word(s) | 8312 | 2021-05-24 03:23:45 | | |
Video Upload Options
Alternative splicing (AS) is a vital post-transcriptional modulator of gene expression that amplifies the proteome diversity and regulates many physiological processes essential for mounting responses to stresses in plants.
1. Introduction
Rice is acclaimed as the most economically important crop serving as a staple food for human consumption worldwide. The rapidly growing global population has profoundly increased the demand for rice, especially in Asian countries [1]. According to the latest report of FAO, global rice production in 2019 was 502.8 million tons (mt); however, the global rice demand in 2020/21 is expected to increase to a record level of 514 mt [2]. Ensuring sufficient rice supply to the growing population has remained a serious challenge for rice breeders, which is exacerbated by the adverse environmental conditions. Environmental stresses severely affect different facets of growth and development and eventually reduce the productivity and yield of rice [3][4]. Therefore, the adverse environmental factors together with the increasing population compel us to develop next-generation climate-smart rice cultivars for sustainable agriculture. However, accomplishing this requires a comprehensive understanding of the complex regulatory mechanisms that contribute to stress tolerance in rice.
Plants have evolved different sophisticated regulatory strategies for adapting to biotic and abiotic stresses [5][6]. Regulation of gene expression at the transcriptional, post-transcriptional, and post-translational levels is crucial for the survival of plants under these stresses [5][7][8]. In recent years, post-transcriptional gene regulation has been increasingly recognized as an important regulatory mechanism in plant growth, development, and especially in stress responses. AS is a pervasive and highly dynamic post-transcriptional regulatory process, which plays a pivotal role in reprogramming gene expression in plants in responses to environmental stresses [9][10], and is gaining attention in the quest to develop stress-tolerant plants. AS enables regulated production of multiple distinct mRNAs and protein variants from a single gene via differential joining or skipping of exons or portions of exons and removal of introns within a pre-mRNA transcript [11]. For general mechanisms of AS, the reader is referred to recent reviews [12][13][14]. Five major types of AS events include intron retention (IR), exon skipping (ES), alternative 5′ splice sites (A5SS; alternative donor site), alternative 3′ splice sites (A3SS; alternative acceptor site), and mutually exclusive exons (MXE) [15][16]. The IR is the most common type of AS event in plants, whereas the ES is the most prevalent in animals [17][18]. The difference in prevalent AS events between plants and animals is likely due to the architectural differences in plant and animal genes, such as the presence of much shorter introns in plants, and also due to the intron-defined splicing process in plants [11][18]. Transcripts with the retained introns are present in polysomes indicating their export to the cytoplasm, and potential regulatory roles in plants [17][19]. Intriguingly, alternative transcripts with retained introns tend to be from genes that function in diverse stress responses [17].
AS is a highly regulated process that requires cis-elements (located in both exonic and intronic sequences of pre-mRNA) and trans-acting factors that bind to these cis-elements—a basic mechanism that is quite conserved in eukaryotes [20][21]. An interplay between these regulatory elements (exonic/intronic splicing enhancers and silencers) and trans-factors determines the fate of a particular exon or intron be retained or excluded from the mature transcript by either facilitating or hampering the assembly of the spliceosome [20][22]. The canonical mechanism of AS suggests that serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) constitute the important trans-factors that bind splicing enhancers and silencers to facilitate and suppress splicing, respectively [16]. SR proteins play crucial roles in both constitutive and AS and regulate the recruitment of splicing machinery to splice sites by mediating the protein-protein interactions among other splicing factors [23].
Besides, increasing evidence indicates that AS is also regulated by the state of chromatin in plants [24]. Due to the co-transcriptional regulation of splicing, it is influenced by the elongation rate of RNA polymerase II, which is in turn controlled by chromatin structure [25]. In fact, the IR events are highly enriched in those genomic regions of plants where chromatin is highly accessible [25]. DNA methylation also regulates AS in rice by affecting the dynamic chromatin structure [26]. The epigenetic regulation of AS is also involved in plant stress responses. For example, modulation of AS through histone methylation serves as a key mechanism for temperature sensing in Arabidopsis [27]. Likewise, the chromatin remodeler ZmCHB101 affects the AS contexts in maize by altering the chromatin and histone modification status, as well as the elongation rate of RNA polymerase II in response to osmotic stress [28]. Moreover, noncoding RNAs have also emerged as key regulators of AS patterns in plants, which, in turn, influence the miRNA-mediated regulation of gene expression associated with different biological processes [29][30][31][32][33][34]. Together, these findings reveal an additional epigenetic layer in regulating AS in plants.
AS is a functional innovation of eukaryotic genomes as it increases the proteome complexity and functional capacity in a cell, and hence, the phenotypic diversity [20][35]. AS of pre-mRNAs has various molecular consequences at the mRNA and protein levels (Reviewed by Laloum et al. [9]). Depending on the different AS events and the alternative inclusion or exclusion of fragments in the mature transcript, AS can produce protein isoforms with distinct domains that may carry different molecular functions. AS-mediated alteration of the open reading frame (ORF) often results in the generation of truncated proteins, due to the introduction of a premature termination codon (PTC) in the mature transcript. The PTC-containing transcripts are then recognized by specific cellular proteins, which mediate the elimination of these transcripts through nonsense-mediated decay (NMD)—a cytoplasmic translation-coupled mechanism that degrades PTC-containing mRNAs [36][37]. Several reports also suggest that PTC-containing mRNAs escape NMD to form truncated translation products with regulatory functions, such as modification of protein interaction networks, negative regulation of protein dimerization, and alteration of post-translational modifications [38]. The truncated isoforms can also act in autoregulatory loops to control the gene transcription [39]. It is also suggested that the production of these truncated proteins serves as a potential way of fine-tuning the amount of functional protein in the cell [40]. AS also occurs in the untranslated regions (UTRs) of pre-mRNAs. The AS in 5′-UTRs is suggested to influence translation efficiency by generating transcript variants with altered upstream ORFs (uORFs) or riboswitches, whereas AS in the 3′-UTR can affect miRNA binding sites in the transcript variants [41].
AS patterns are highly altered in response to different environmental stresses, which allows plants to rapidly adjust the abundance and function of crucial stress-responsive proteins [9][10]. Comparative plant genome analysis has shown that genes undergoing AS have a greater evolutionary rate, which may be associated with better adaptation to adverse environmental conditions [38]. Many next-generation sequencing (NGS) studies have revealed that a considerable number of AS events occur in plants in response to environmental stresses [10][42]. Global transcriptomic studies using RNA-seq have revealed that stress-induced AS occurs mainly in regulatory genes, such as transcription factors, protein kinases, and splicing factors [10][42][43]. Because most of the stress-induced AS patterns are discovered using transcriptomic methods, the functional significance of a majority of the stress-regulated AS events in plants is yet to be demonstrated. However, increasing evidence reveals that AS in individual genes plays critical functional roles in various aspects of plant stress responses [9][10][44][45]. Besides, emerging evidence indicates that AS is involved in an intriguing adaptive response of stress-memory in plants. Heat stress priming-induced AS memory has been observed in Arabidopsis plants, which helps them survive subsequent lethal stress by generating appropriate stress-responsive splice isoforms [46].
As in other plants, AS is a key regulatory mechanism that is modulated in response to various biotic and abiotic stresses in rice also. As discussed below, RNA-seq has been widely used in rice to analyze AS in response to various abiotic (including drought, salinity, extreme temperatures, light stress, hypoxia, metal stress, mineral stress, and abscisic acid) and biotic stresses (including bacteria, fungi, insects). In addition to the transcriptomic studies, there are several studies on AS in individual candidate genes in rice with functional significance for stress tolerance.
2. Abiotic Stress
2.1. Transcriptomic Analyses of Abiotic Stress-Induced AS
High-throughput sequencing technologies have become indispensable to study epigenomics, genomics, and transcriptomics. In particular, RNA-seq is a powerful tool that has truly revolutionized our ability to analyze plant transcriptomes, especially as it relates to environmental stresses. RNA-seq has catalyzed plant science research by enabling the rapid profiling and detailed investigation of transcriptional and post-transcriptional processes in any plant with or without a sequenced genome [24][47]. RNA-seq has rapidly superseded other transcriptomic methods for studying gene expression dynamics as it offers many advantages, such as detection of transcripts beyond the limit of microarrays, increased dynamic range for more extreme limits of detection, and the ability to identify novel, rare, low-abundance, and alternatively spliced transcripts in a single sequencing run [24][47][48]. It can detect not only the differential gene expression, but also the differential AS events in different genetic backgrounds under a particular condition [11]. Besides, RNA-seq can robustly detect single nucleotides polymorphisms (SNPs) [49].
RNA-seq studies in rice have provided new perspectives on unknown transcript dynamics and led to significant progress in identifying genes and molecular mechanisms that regulate important traits governing abiotic stress tolerance [50][51][52][53][54][55]. These studies have revealed altered patterns of splicing in key genes associated with various abiotic stress responses. Below, we discuss rice transcriptome studies that are aimed at understanding AS in rice in response to various abiotic stress conditions.
2.1.1. Relationship between Stress-Induced AS and Stress-Tolerance
The transcriptome sequencing is performed on the genotypes with inherent contrasting genetic responses to stresses. This is done to unravel the key molecular components that confer differential stress tolerance in the contrasting genotypes. Similarly, RNA-seq has provided novel insights into differential AS in contrasting rice genotypes under different abiotic stresses, which might offer new avenues for rice breeding. A transcriptomic study revealed divergence in drought-induced AS patterns between upland drought-tolerant and lowland drought-sensitive rice cultivars, with more cultivar-specific AS events in tolerant than sensitive cultivar [56]. Spliceosome-associated genes encoding hnRNP G, SR, and RNA helicase proteins were largely subjected to AS in upland cultivar, and this distinctive AS in spliceosomal proteins may lead to the differential AS in the other genes. In fact, genes associated with stress-associated pathways, such as circadian rhythm and oxidative phosphorylation, showed AS patterns specifically in upland rice. Interestingly, several genes encoding spliceosomal proteins with different AS events were located in drought-responsive quantitative trait loci (QTLs), implicating a role for these genes in rice response to drought. These results indicate that AS of pre-mRNAs encoding splicing factors can be a crucial strategy adopted by upland rice during long domestication to tolerate drought. Another transcriptomic study associated with salt stress in contrasting rice genotypes has revealed down-regulation of AS, specifically in sensitive genotype, which might contribute to its susceptibility to salt stress [54]. A systematic comparison of transcriptomic profiles of contrasting rice genotypes under salt stress has uncovered AS to be a crucial mechanism underlying salt tolerance in salt-tolerant genotype FL478 [57]. Although more AS events were identified in susceptible genotype than the tolerant, AS events in genes involved in crucial salt tolerance-associated processes of ion-transport and signaling pathways were more frequent in tolerant genotype. These alternatively spliced variants were either highly expressed (bZIP, PP2C variants) or specifically expressed (WRKY30, MAPK kinase, and HAK25, ABC and ZIFL transporter variants) in the tolerant genotype [57]. Similar results were reported in the case of salt-tolerant barley and susceptible rice genotypes under salt stress. Although more AS events were found in rice than barley, a higher level of AS in the barley genes related to ion-transporters and transcription factors contribute to the higher salt tolerance in barley, due to the superior K+/Na+ homeostasis [58]. Moreover, one of the SR splicing factors was specifically expressed in barley. Likewise, a separate transcriptomic analysis revealed higher AS events in salt-sensitive than salt-tolerant genotype, but a higher number of alternatively spliced isoforms were specifically expressed in tolerant genotype as compared to sensitive genotype under salt stress [59]. In this study, although most of the AS events decreased in all the analyzed rice genotypes, including a drought-tolerant genotype under desiccation stress, high expression of some spliced transcripts was observed exclusively in the drought-tolerant genotype, which might contribute to drought tolerance. Alkali stress response in contrasting rice cultivars revealed more AS events in alkali sensitive cultivar; however, some alkali-responsive genes were differentially alternatively spliced between the two genotypes, suggesting a potential role for AS in the differential response of the two cultivars to alkali stress [52]. In another study, a lower number of AS events were found in an iron stress-tolerant rice genotype under iron-excess stress than control [60]. However, among the different splicing events, IR was more prevalent, and the splicing events were altered in an organ-specific manner under stress.
Collectively, the above five studies indicate that higher stress-induced AS in a genotype does not necessarily correlate with stress tolerance, but the frequency of genotype-specific events, the abundance of AS transcripts related to key stress-responsive processes, and the nature of splicing events are likely to contribute to stress tolerance. It is possible that the bulk of alternatively spliced transcripts may be subjected to degradation by NMD and may not be translated. In fact, only about 14% of the total alternatively spliced transcripts are translated into peptides under hypoxic stress in rice [61]. In a related study, treatment with a salt-tolerant endophyte was shown to confer salt stress adaptation and decrease the AS in salt-sensitive rice plants as compared to their untreated counterparts [62]. It was proposed that treatment with the endophyte causes a lowered perception of salt stress signaling, which might not activate the stress-responsive mechanism of AS in the treated plants, underscoring the importance of AS in response to stress. Another RNA-seq analysis at the reproductive stage of a drought-resistant rice maintainer line and its recurrent parent has revealed a similar level of gene expression at the transcriptional level under drought; however, the increased level of AS in transcription regulatory networks likely contributes to the genetic basis of adaptation to drought in the maintainer line [51]. It was reported that differentially expressed genes (DEGs) involved in the biological process of chloroplast components undergo increased AS in a maintainer line and are regulated by six transcription factor families, adding an extra regulatory level to genetic improvement of drought resistance in this line. The higher drought resistance in offspring could be due to AS-mediated limited damage to the photosynthetic apparatus in chloroplasts for maintaining the energy metabolism under drought. Li et al. [63] reported that changes at the transcriptional level do not always correspond to the changes in protein abundance. They have shown that in salt-tolerant genotype, the number of up-regulated DEGs was markedly higher at the translational level as compared to transcriptional level, whereas the number of up-regulated DEGs in susceptible genotype was comparable at the two levels. However, the number of up-regulated DEGs in the susceptible genotype was lower at the translational level as compared to salt-tolerant genotype, which could be attributed to the almost 3-fold abundance of alternatively spliced transcripts in tolerant genotype as compared to susceptible genotype that could increase proteome diversity in tolerant genotype under salt stress.
2.1.2. AS-Mediated Spatial Regulation of Stress-Responses
Rice responds to abiotic stresses by spatially activating distinct molecular processes [64][65]. Transcriptomic analyses have revealed that AS may play key roles in regulating the stress-induced processes in roots and shoots of rice. A high level of AS occurs in root and shoot of rice under aerobic conditions [53], with more splicing events occurring in the aerobically-induced chloroplast development-associated genes (coding for tetratricopeptide repeat domain-containing protein and GOLDEN2-LIKE1 transcription factor) in the shoots; and higher splicing events taking place in the aerobically-induced plastid-related gene (coding for PAP fibrillin domain-containing protein) in the roots. Similarly, in wild rice, Oryza longistaminata, the number of genes undergoing AS and the frequency of splicing events greatly increase in shoots and rhizomes under chilling stress. In shoots, AS occurs exclusively in the chilling-induced genes associated with regulation of gene expression and photosynthesis; whereas, in rhizomes, the chilling-induced genes associated with Ca2+-binding undergo AS [66]. It indicates that in response to chilling, AS may regulate calcium-mediated signal transduction in rhizomes, whereas it may play crucial roles in regulating transcription factors having diverse functions and also the photosynthesis in shoots. Therefore, organ-specific AS may contribute to the regulation of long-term chilling tolerance in O. longistaminata. In both of these studies, a higher number of AS events were observed in the shoots than in the roots, and vice-versa for the number of DEGs. In response to stress, the DEGs involved mostly in signaling pathways (including hormone signaling) are activated in roots, which may send stress signals to shoots where they may induce higher AS of certain genes for stress adaptation by regulating the expression of AS-related factors. Wild rice species, which are generally tolerant to various environmental stresses, appear to reap the benefits of AS for abiotic stress tolerance differently. Under phosphorus stress, O. rufipogon adopts a distinct AS-based strategy than the one used by O. longistaminata under cold stress. In O. rufipogon, phosphorus stress causes up-regulation of Lsm8 and U1A spliceosome-related proteins and down-regulation of SPF and SR splicing factors [67]. It was proposed that up-regulation of Lsm8 and U1A may increase the AS, while the down-regulation of the two splicing factors may lead to the changes in pre-mRNA splice sites under phosphorus stress, which may be associated with the phosphorus stress tolerance in this wild rice species. The use of increased AS by O. rufipogon as a strategy to deal with the phosphorus stress was also supported by the finding that phosphorus stress reduces the level of transcription-related genes and increases the RNA splicing and translation elongation, indicating that phosphorus-induced gene expression operates at the post-transcriptional level.
2.1.3. Stress-Induced Changes in AS Represent an Independent Layer of Gene Regulation
In addition to the DEGs, AS also occurs in genes that are not affected at the transcription level between the samples (called differentially alternatively spliced genes; DASGs) under stress. In other words, AS can also be independent of transcriptional regulation, which indicates that AS serves as an additional layer of regulation [11]. In this regard, almost 1741 DAS events were identified in rice seeds under hypoxia, out of which, more than 95% did not overlap with DEGs, suggesting that DASGs is a different group of genes than that of DEGs that are responsive to hypoxic germination, suggesting that AS may have a distinct role in the germination of rice seeds under hypoxia [61]. Intriguingly, the DEGs and DASGs are functionally enriched in distinct cellular processes and metabolic pathways, with cellular metabolism and cell growth mainly enriched in DEG data; while protein degradation, post-transcriptional regulation, and transport processes mainly enriched in DASG data sets. This study also found that just 38% of hypoxia-induced DAS events were translated into peptides in the germinating seeds, indicating that most of the DAS events may be degraded via NMD. Similar results for the nonoverlap between DEGs and DASGs and their involvement in functionally distinct processes have been obtained in rice under mineral stress [11], drought [68], and cadmium stress [50]. It is inferred from these studies that regulation of stress-induced gene expression at transcriptional and post-transcriptional (AS) levels is independent, supporting the notion that environmental stresses influence the gene expression at transcriptional and AS levels [21]. These two mechanisms act in concert to promote the performance of rice under stress conditions; e.g., high transcriptional expression of cellular metabolism-related genes can lead to significant misfolded protein production under hypoxia, whereas the AS-mediated generation of new isoforms may potentially degrade these misfolded proteins to alleviate the hypoxic stress [61]. This suggests that adaptation of rice to stresses may have been shaped by distinct transcriptional rewiring of AS, as well as by gene expression variation.
The transcriptomic studies also reveal that SR splicing factors themselves undergo extensive DAS, without exhibiting differential expression, under abiotic stresses [11][50][61] as a means to regulate the AS of more diverse downstream genes. In fact, Chen et al. [61] report that hypoxia-induced AS in SR proteins can generate diverse isoforms of these splicing factors with a different choice of splice sites, and this was associated with the abundance and preferential usage of certain noncanonical 5′-splice sites for germination under hypoxia.
2.2. Abiotic Stress-Induced AS of Candidate Genes
2.2.1. Abiotic Stress-Responsive Genes
Although global transcriptome analyses have identified plenty of abiotic stress-induced alternatively spliced transcripts, the functional significance of only some of them in adaptive stress responses has been demonstrated. The protein-coding genes can be either structural or regulatory, with the proteins encoded by structural genes having structural or functional properties in the cell—while those encoded by regulatory genes regulate the expression of structural genes [69]. Based on our extensive literature survey, we found that rice genes encoding regulatory proteins are more likely to undergo AS under various abiotic stresses, and more importantly, the functional significance of AS events in these genes has been analyzed (Figure 1; [70][71][72][73][74][75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95]). It follows that AS is an elegant and energy-saving mechanism particularly targeting those genes which can, in turn, regulate a diverse set of downstream structural genes for activating the adaptive responses to stresses. However, some structural genes of rice also undergo stress-induced AS, which has important implications for rice abiotic stress tolerance. AS in abiotic-stress-responsive regulatory and structural genes of rice acts through diverse mechanisms as illustrated below.
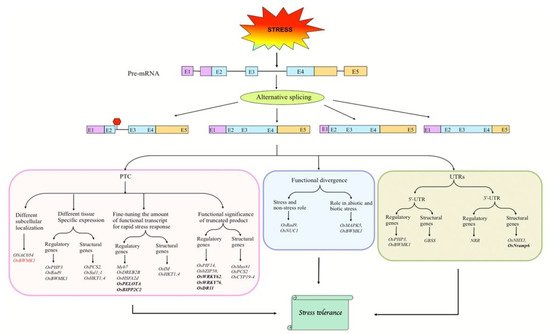
Figure 1. Various modes of AS-dependent regulation of splice isoforms in response to biotic and abiotic stresses. Under stress conditions, the pre-mRNA of stress-responsive genes may generate alternative transcripts with PTC that may regulate certain features of the splice isoforms, such as subcellular localization and spatial expression, which may fine-tune the level of functional protein isoforms or produce proteins with altered functions (Left). The alternative transcripts may be functionally distinct (Middle). Besides the coding region, stress-induced AS in UTRs of pre-mRNAs can also regulate splice isoforms (Right). Genes in bold represent biotic stress-responsive genes, and the remaining ones are abiotic stress-responsive. Gene shown in red functions in both biotic and abiotic stresses. Boxes and lines represent exons and introns, respectively. In transcripts, purple, sky blue, and orange colors represent 5′-UTR, coding region, and 3′-UTR, respectively. PTC, premature termination codon; Red hexagon represents PTC.
One of the most common mechanisms by which AS can regulate the transcript abundance in response to stress is via the cytoplasmic RNA degradation system of NMD. Transcripts that are particularly prone to NMD have classical features of retained introns, PTC, uORFs, etc. [15]. However, these transcripts may also escape NMD to form truncated proteins with key regulatory functions under stress [9][20][38].
Fine-Tuning the Abundance of Functional Transcripts
By producing truncated nonfunctional proteins, AS through timely IR or PTC serves as a potential way of fine-tuning the amount of stress-responsive functional protein in the cell during abiotic stress. For example, drought and heat stress-induced AS regulates the cellular levels of OsDREB2B, which has a nonfunctional (OsDREB2B1) and a functional transcript form (OsDREB2B2). The nonfunctional and functional forms are predominant under non-stress and stress conditions, respectively. Under drought, the functional form operates to increase drought tolerance by enhancing the plant survival rate and relative water content (Figure 2A) [70]. Similarly, hypoxia-induced myb7 has an unspliced (with two retained introns) and a spliced form, and the ratio of these isoforms changes under aerobic and hypoxic conditions. The spliced form increases the expression of myb-related genes in hypoxic roots [71]. Likewise, the heat shock factor gene OsHSFA2d constitutively produces the inactive forms (OsHSFA2dII and OsHSFA2dIII) under non-stress conditions, whereas under heat stress, it generates the transcriptionally active form (OsHSFA2dI), which participates in heat stress response by regulating the unfolded protein response (UPR) (Figure 2B) [72].
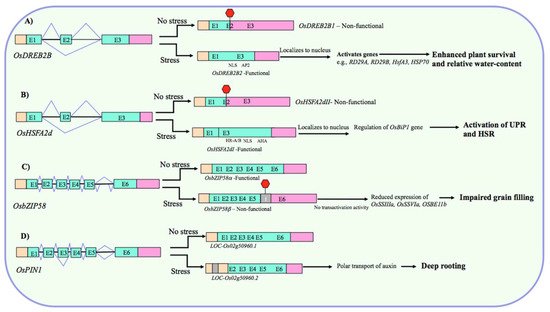
Figure 2. Representative examples of AS-mediated regulation of gene expression in rice under heat/drought stress conditions. (A) Under nonstress conditions, nonfunctional (due to frameshift and PTC) form predominates, while the functional form (with intact NLS and typical AP2/ERF domain) is produced under drought to increase drought tolerance potentially by enhancing the expression of stress-related genes [70]. (B) Under nonstress conditions, nonfunctional (due to PTC) form predominates, while the functional form (with intact DNA-binding-, oligomerization-, and activation-domains) is produced under heat stress which participated in heat stress response (HSR) by regulating the expression of unfolded protein response (UPR) marker gene OsBip1 [72]. (C) Under nonstress conditions, functional form predominates, while the nonfunctional form (due to IR-mediated PTC), lacking bZIP domain, is produced predominantly under high temperature, which causes impaired grain filling by repressing the expression of genes associated with grain filling [76]. (D) An IR event in the OsPIN1 transcript has a potential functional significance in regulating the rice deep rooting under drought by facilitating the polar auxin transport [68]. Boxes and lines represent exons and introns, respectively. In transcripts, peach, aquamarine, and violet colors represent 5′-UTR, coding region, and 3′-UTR, respectively. Red hexagons represent PTC. “I” in the splice isoforms indicates an intron retention event.
Through IR and PTC, AS also regulates the abundance of functional stress-responsive transcripts of some important rice structural genes. The best example of this is provided by the OsHKT1;4-mediated Na+ exclusion from the leaf blades of salt-stressed rice [73]. OsHKT1;4 has three transcript variants, among which, only the canonically spliced one is functional. The high Na+ excluding pokkali is superior to salt-sensitive Nipponbare in minimizing the Na+ load in the young photosynthetic leaf blades by maintaining a high ratio of functional OsHKT1;4 transcript variants in the corresponding sheaths. Similarly, the pre-mRNA of an oxidase gene OsIM produces functional (OsIM1) and pseudo-transcript (OsIM2), and both exist under non-stressed conditions; however, under salt stress, the OsIM1/OsIM2 ratio greatly increases because of the significantly reduced OsIM2 levels [74], that may relieve salinity-induced oxidative stress. Likewise, the iron-superoxide dismutase gene OsFe-SOD has two isoforms, OsFe-SODa and intron-containing OsFe-SODb, which respectively show significantly high and low expression under cold stress [96].
Overall, these studies indicate that an inactive transcript variant may exist to keep its gene constitutively active without affecting the plant growth, while the onset of stress may send a signal to the splicing apparatus to alter the splicing pattern to produce the active stress-responsive form. The rationale behind this elegant ON/OFF switch is that plants can promptly generate the active transcript forms as an adaptive strategy when subjected to abiotic stress, and by doing so, they save time that is otherwise needed for transcription and transcript accumulation of the gene under discussion [70].
Regulatory Role of IR in Stress Responses
Previously, limited attention was paid to IR primarily because it was assumed to be a product of missplicing with no physiological implications, but diminishing the gene expression by NMD. However, accumulating evidence has revealed that it acts as a critical component of the controlled gene expression program and plant abiotic stress responses [45]. The case studies of individual IR events have demonstrated it as an exemplar of regulated splicing and have provided key mechanistic insights into regulating abiotic stress responses in rice. For example, cold-induced AS in the pre-mRNA of phytochrome-interacting factor OsPIF14, which regulates cold tolerance in rice by repressing the expression of OsDREB1B, introduces a PTC via intron-retention, which leads to the production of a transcriptionally inactive truncated protein lacking a complete bHLH domain [75]. At severe temperature, the inactive form OsPIF14β is up-regulated and the active form OsPIF14α is down-regulated. This has a functional significance because OsDREB2B is crucial for rice cold tolerance and its activation (due to down-/up-regulation of α-/β-form) at lower temperature leads to the accumulation of OsDREB2B, and subsequent cold tolerance. The IR in another transcription factor gene OsbZIP58 has an important role in mediating the effect of high temperature on rice grain filling and seed quality control [76]. The high temperature triggers AS of OsbZIP58 to generate mainly the intron-retaining truncated form OsbZIP58β over the full-length form OsbZIP58α (Figure 2C). The predominance of transcriptionally less active OsbZIP58β at high temperature impairs the accumulation of storage materials in rice grains possibly by diminishing the amount of functional OsbZIP58 protein, leading to the reduced trans-activation of genes associated with the grain filling, such as OsSSIIIa, OsSSVIa, OsBEIIb, GBSSI, and OsBEI (Figure 2C). Intriguingly, this IR event is also associated with the heat-sensitivity of contrasting rice genotypes [76], suggesting that it can potentially act as a heat tolerance marker in rice. Another example of a functional AS event comes from an auxin efflux carrier gene OsPIN1 [68]. Dynamic AS is associated with deep rooting-mediated drought avoidance in rice and overexpression of OsPIN1 containing a retained intron improves the ratio of deep rooting (RDR) in rice (Figure 2D). Because OsPIN1 gene and the spliced isoform did not show any differential expression between low and high RDR rice genotypes and because this IR event was highly up-regulated in deep roots of high RDR genotypes only, it is likely that IR event in OsPIN1 is important for rice root development and increases RDR apparently by facilitating the auxin transport [68]. In yet another case, the unspliced form of rice myb7 having two retained introns and a short leucine zipper is predominant in the aerobic organs; while the introns are spliced out under hypoxic, suggesting a regulatory role of this intron as an efficient switch to alternatively express myb regulatory protein under contrasting conditions [71]. Alternatively, if we analyze this finding from the perspective of aerobic conditions being stressful to rice [47], then this IR as a functional event makes sense because the leucine zipper motif encoded by unspliced intron can potentially interact with the myb factor [97] to activate the transcription of the target genes under aerobic stresses.
Small sequence variations in certain genes can have a great impact on their AS and expression. For example, insertion of a mutation in the post-translational modification-related glycosylphosphatidylinositol gene OsGPI8 causes IR, PTC, and loss of function, especially at elevated temperature [98]. The IR-mediated silencing of OsGPI8 impairs cell wall biosynthesis, cell division, and cell shape in rice plants, leading to their fragility and drooping of shoots. This finding indicates a potential interplay between the processes of post-transcriptional and post-translational gene regulation for eliciting the proper abiotic stress responses in rice.
Analyses of IR in structural genes have also revealed prominent functional roles in biological settings for rice abiotic stress tolerance. The DNA repair gene OsMus81 produces two splice variants (the main OsMus81α and the truncated OsMus81β), which are up-regulated under intense light, UV-C, and γ-radiation [77]. Because OsMus81β lacks the helix-hairpin-helix motif, only OsMus81α interacts with Mms4/Eme1 to form a hetero-trimeric endonuclease complex, which functions in nucleotide excision repair. Besides, both of them interact with recombination repair protein OsRad54.However, because OsRad54-OsMus81β interaction is stronger than that of OsRad54-OsMus81α, OsMus81β may operate at the later stages of homologous recombination pathway for DNA repair [77]. This indicates that these alternative transcripts may repair the DNA damage by different repair mechanisms. Similarly, the phytochelatin synthase gene OsPCS2 has a canonically spliced variant (OsPCS2a) and the alternatively spliced PTC-containing variant (OsPCS2b). These isoforms are differentially expressed in cadmium-stressed roots and shoots. It was suggested that the unproductive OsPCS2b splice variant has a regulatory role in controlling the tissue-specific expression of the functional variant during cadmium stress [99]. Further, Oryza species-specific sequence distinctions in the vacuolar NHX-type antiporter (OsNHX1) relate to the differential intron-retention at the 5′ and 3′ ends of its transcript [100]. The recent IR event in 5′-UTR is found only in AA Oryza genomes, while the retention of the 13th intron is more ancient in origin and also occurs in OcNHX1 of halophytic wild rice (O. coarctata). Interestingly, species- and tissue-specific switches in the expression of these splice variants occur under salt stress, indicating that IR may contribute to the differential ability of Oryza NHX1 homologs to maintain ion-homeostasis under salt stress. IR in cold-responsive OsCYP19-4 transcripts provides another example of a functional effect of AS [78]. The alternative transcripts OsCYP19-4.2/3 are truncated, but their interaction with AtRCN1 (regulator of auxin signaling in guard cells) indicates their role in cold acclimation by influencing stomatal physiology. Besides, most of these truncated isoforms show different subcellular localization under cold stress, indicating their organelle-specific roles [78].
From these studies, it is obvious that intron-retaining mRNAs are pivotal for regulating normal plant physiology. Therefore, IR should no longer be perceived as a selfish burden on the nucleus that only reduces gene expression with no functional significance in plant biology. In nutshell, the IR can be a robust mechanism to regulate gene expression with potential functional implications for rice stress tolerance.
Tissue- and/or Developmental Stage-Specific Expression of Transcript Variants
Consistent with the results from genome-wide studies, the expression of abiotic stress-induced individual mRNA variants of several regulatory and structural genes is development- or tissue-specific [73][79][83][99][101][102], while the variants from other genes show differential expression in various tissues and/or at developmental stages [77][81][96][102][103][104][105][106][107][108]. The spatial expression of splice variants has a potential significance for rice stress tolerance. For example, high expression of the functional form of OsHKT1;4 in young rice sheaths causes their loading with Na+ ions to protect the young photosynthetic leaf blades from Na+ toxicity, while the lower levels of the functional form (due to higher expression of truncated forms OsHKT1;4-SV1/SV2) in the older sheaths let Na+ ions go through to the older blades for storage without harming the plant [73]. The expression of light-regulated AS transcripts of rice pseudo-histidine phosphotransfer protein 3 (OsPHP3) is differentially modulated by auxin in rice roots, causing high root-specific expression of OsPHP3.2/3.3 isoforms [83]; indicating that auxin may mediate the effect of light signaling on root growth by differentially regulating the abundance of AS transcripts of OsPHP3. Similarly, the higher ratio of functional to nonfunctional AS forms of OsPCS2 (OsPCS2a/OsPCS2b) in the cadmium-stressed shoot tissue of rice potentially reduces the cadmium toxicity in shoots [99]. Likewise, the AS variants of cold-responsive OsFe-SOD show differential expression at different developmental stages of rice, with the expression of OsFe-SODb being higher at the vegetative stage, while that of OsFe-SODa being higher at the flowering stage [96]. Altogether, these results support the notion that tissue-specific AS events may play key roles in fine-tuning molecular and physiological processes in response to abiotic stress by regulating the relative abundance and function of the encoded proteins. They also signify that AS events may be important for mounting response to stress at particular developmental stages.
Stress and Non-Stress Roles of Alternative Transcripts
A transcript variant produced from a particular regulatory gene may be responsive to abiotic stress, whereas its other variant(s) may not be stress-responsive, but could be involved in other processes instead. For example, OsRad9, involved in the cell cycle check-point signaling pathway, generates transcript variants of OsRad9.1 and OsRad9.2. OsRad9.1 is the full length and main cell cycle checkpoint protein involved in rice responses to different abiotic stresses; while the shorter OsRad9.2, having lost nine phosphorylation sites at its C-terminal, is highly expressed in pollen [79]. Since phosphorylation serves as a key modification in signaling pathways, the loss of phosphorylation sites in the shorter variant may have altered its function, suggesting that AS may expand the repertoire of functions of OsRad9—the usual role in the repair of DNA damage and a different role in pollen development. Similarly, the nucleolin gene, OsNUC1, generates two isoforms, the longer OsNUC1-L is involved in the root growth, and the shorter OsNUC1-S with no N-terminus is involved in salt tolerance as determined by the phenotypes of the OsNUC1-L- and OsNUC1-S-transgenic plants, respectively. It is proposed that the deletion of the N-terminal makes the shorter isoform function as an mRNA stabilizer for the other salt-tolerant genes under salt stress [80]. Similarly, OsMAPK5 produces longer abiotic stress-induced OsMAPK5a with intact kinase activity and shorter OsMAPKb lacking this activity, suggesting AS regulation of this gene in response to stress [109].
Regulation of Subcellular Localization of Transcript Variants
The decision of whether an RNA is translated, modified, preserved, or degraded, is dictated by the intracellular location of the RNA, which in turn determines the biological function of the RNA [110]. Subcellular targeting can, therefore, have a significant influence on the regulatory and biochemical potential of the translated protein. AS influences the localization of spliced variants of some abiotic stress-associated genes of rice, thus regulating the function of their translation products. The rice senescence-related ONAC054 mRNA produces ONAC054α which encodes an endoplasmic reticulum (ER) membrane-bound protein during normal conditions; however, the stress hormone abscisic acid (ABA) induces AS of ONAC054 transcript to generate the truncated transmembrane domain (TMD)-less ONAC054β [82]. ONAC054β relocates from ER to nucleus, where it activates ABA signaling and senescence-related genes. Similarly, the rice ER stress regulator OsbZIP74 is encoded with an intact TMD under non-stress conditions; however, heat and ER stresses cause this domain to be spliced out from the mRNA of OsbZIP74, and the protein from this splice isoform is then relocated to the nucleus to promote the UPR-related gene expression, thus mitigating the heat and ER stress by enhancing the protein folding [111]. These results imply that the activity of TMD-containing transcription factors depends on their nuclear localization and TMD cleavage, which are in turn regulated by AS. In another case, alternative splice variants of a kinase gene, OsBWMK1, also exhibit differential subcellular localization under oxidative stress, with the larger isoform (OsBWMK1L) imported to the nucleus, but the localization of the smaller and medium variants (lacking the N-terminal sequence present in a larger variant) remaining the same [81]. Notably, salt and oxidative stresses up-regulate the expression of shorter and medium isoforms, but the expression of longer isoform does not change; instead, it responds to oxidative stress at the AS level by relocating to the nucleus and possibly activating the stress-responsive gene expression, indicating the functional significance of this AS event. It implies that OsBWMK1 isoforms act through distinct regulatory mechanisms in response to abiotic stress. Therefore, AS-mediated nuclear import of different proteins occurs via distinct mechanisms—in some cases, it may be a cause of cleavage of a sequence from the imported protein, and in other cases, it may cause retention of a sequence needed for protein import.
AS in UTRs of Pre-mRNAs
Besides the introns and exons in the coding region, AS also occurs in UTRs of pre-mRNAs [41]. One of the AS mechanisms used by most eukaryotes to increase transcript diversity of a gene is the use of alternative first exons by alternative promoter usage. In other words, AS occurring upstream of the first exon of pre-mRNA can result in the usage of alternative promoters by the transcript variants [112]. Thus, such transcript variants differ only in their 5′-UTRs. This mechanism substantiates the results obtained by Koo et al. [81], as discussed above. The differential expression of the three OsBWMK1 transcript variants under stress results from AS and the alternative promoter usage [101]. The constitutive expression of larger isoform (OsBWMK1L) is due to the use of promoter-containing TATA-rich region, while the stress-induced expression of smaller isoform (OsBWMK1S) is due to the use of alternative promoter harboring several stress-responsive cis-elements. In a similar study, the transcript variants of OsPHP3, produced via alternative promoter usage, show differential expression in response to a light signal, with OsPHP3.1 being the major transcript variant in the light-grown seedlings and OsPHP3.2/3.3 predominating in the etiolated seedlings [83]. Notably, the analysis of the upstream sequence of OsPHP3.2/3.3 pre-mRNA, which is spliced out in the case of OsPHP3.1, showed the presence of several light-related cis-elements, including nine activating sequence factor-2 (ASF2)-binding sequences, which were suggested to be highly activated under darkness. Thus, the alternative promoter usage for the generation of transcript isoforms provides an extra route to creating novel regulatory opportunities under stressful conditions.
AS in 5′-UTRs can influence the efficiency of gene expression by generating transcript variants with altered uORFs [41]. It is reported that a uORF can either induce mRNA degradation via NMD or repress the translation of the main downstream ORF [41]. In the case of rice, the functional relevance of a uORF within the 5′-UTR of Nitrogen Limitation Adaptation 1 (OsNLA1) has been demonstrated for the regulation of phosphate acquisition in roots [84]. This gene produces AS variants having long (OsNLA1.1) and short (OsNLA1.2/1.3) 5′-UTRs, with only OsNLA1.1 containing a uORF of 30 amino acids. Among the three isoforms, the longer uORF-containing isoform is the most abundant and exhibits the highest expression in response to high phosphate stress in roots and shoots. Of note, unlike its homolog in Arabidopsis, OsNLA1 is not regulated by the phosphate starvation-induced miR827 [113]. Instead, the uORF and other promoter elements were found to be cooperatively regulating the expression of OsNLA1 for controlling the phosphate uptake and translocation [84]. Importantly, at different phosphate concentrations, uORF-mediated regulation of OsNLA1 expression might occur through mechanisms, such as ribosome stalling, to induce the expression of NLA1.1 at high and repress its expression at low phosphate concentration [114]. Hence, AS in the 5′-UTR plays a key role in phosphate stress tolerance by retaining a regulatory uORF in the most abundant isoform OsNLA1.1 and substituting for the miRNA-mediated regulation. AS in 3′-UTRs can also produce variants with different stress-responsive roles. Rice Nutrition Response and Root growth (NRR) pre-mRNA undergoes AS in the 3′-UTR to produce a longer NRRa and a shorter NRRb transcript, which show varied expression patterns in roots and shoots under macronutrient deficiency, with the larger isoform playing a more crucial role in root growth [85]. Since the silencing of both isoforms results in more improved root growth than when either of them was suppressed, it indicates that these isoforms may act in concert under nutrient stress to modulate the rice root architecture. Therefore, the generation of transcript isoforms via AS in 3′-UTR may provide a means to facilitate the synthesis of functionally related proteins that cooperatively mediate a certain biological response. Likewise, the transcript of OsCPK17 produces five splice variants with most of them down-regulated immediately under the cold stress; however, the expression of isoform OsCPK17.5, having no 5′-UTR and different 3′-UTR as compared to the other isoforms, changes considerably only after 72 h of stress, indicating that this isoform may be involved in cold tolerance during prolonged stress which is controlled by AS [108].
AS in the UTRs of structural genes also has important implications for rice abiotic stress tolerance. The amylose content in rice grains is temperature sensitive and depends on the 5′ splice sites of intron present in the leader sequence of granule bound starch synthase (GBSS)-encoding waxy gene [86]. Differential temperature-dependent utilization of leader intron splice sites results in the generation of GBSS transcripts with dissimilar 5′-UTR sequences. A splice site used at lower temperature causes a large deletion in the 5′-UTR sequence and also generates a uORF, which does not prevent the accumulation of mature GBSS mRNA. However, the splice site used at higher temperatures causes the production of a substantial amount of incompletely processed mRNA and also generates a uORF, which selectively degrades GBSS transcript [86]. The importance of UTRs in rice salt tolerance is presented by AS in the 3′-UTR of OsNHX1, which has three transcript variants—one of which has a truncated C-terminal end with a deletion of 150 bp in the 3′-UTR. Transgenic rice plants overexpressing the transcripts with intact 5′- and 3′-UTRs exhibit higher salt tolerance—due to significantly more K+ level and chlorophyll-retention at seeding stage and more spikelet fertility and yield at the reproductive stage—than those expressing the 3′-UTR-truncated transcript [87]. Because many of the rice stress-related genes undergo AS, the study underscores the importance of using full-length transcripts for generating more stress-tolerant transgenic plants.
AS and ABA-Mediated Responses
Phytohormone ABA, which is involved in the adaptive abiotic stress responses by controlling the expression of various stress-responsive genes, plays a crucial role in the AS-mediated regulation of rice abiotic stress responses. AS can target the components of ABA signaling to fine-tune the stress responses. For example, the transcript variants of OsABI5 perform distinct functions in ABA signaling by differently interacting with ABA signaling protein OsVP1 and distinctly binding the stress-responsive ABA-related G-box element [115]. ABA also regulates key cellular processes in rice by controlling the AS of specific genes. ABA-mediated AS in the mRNA of ER-resident ONAC054 produces a transcript variant (ONAC054β) that is translocated to the nucleus for activating the genes related to senescence and ABA signaling [82]. Moreover, the transcript isoforms of SUMOylation-related gene SUMO-conjugating enzyme 1a (OsSCE1a) are differentially expressed in response to ABA [116], indicating that AS may fine-tune the regulatory effect of ABA on SUMOylation machinery. Further, although the functional transcript variant of OsIM (OsIM1) is induced by ABA, the high ABA concentrations repress it, indicating a potential feedback inhibition, and hence, the involvement of OsIM1 in the ABA-biosynthesis [74]. The study also suggests that because ABA is the end-product of the carotenoid biosynthesis pathway, OsIM1 can be involved in rice response to ABA by promoting carotenoid biosynthesis. Similarly, OsMAPK5 has two splice isoforms, OsMAPK5a and OsMAPK5b. In contrast to OsMAPK5b, OsMAPK5a has kinase activity and is ABA-inducible. The OsMAPK5a-transgenic plants show increased drought, salt, and cold tolerance [109]. Along similar lines, alternatively spliced variants of OsGATA23 are differentially regulated in salt-tolerant and sensitive rice genotypes in response to ABA and other abiotic stresses [107]. Collectively, these studies indicate that AS may adjust the effect of ABA on rice abiotic stress responses and that ABA mediates the post-transcriptional regulation of rice stress responses. In fact, some splicing regulators of rice (described below) show altered response to ABA [117][118][119]), implying that ABA may be involved in the control of rice abiotic stress responses by these splicing factors.
Regulation of AS by Abiotic Stress-Responsive Genes
In addition to the above-mentioned studies, other studies on rice abiotic stress-responsive genes have revealed that they are not themselves alternatively spliced but influence the stress tolerance of rice by regulating the splicing of other genes. For example, the rice immunophilin family gene OsFKBP20-1b participates in drought and salt stress responses by positively affecting transcription and pre-mRNA splicing of abiotic stress-related genes. OsFKBP20-1b-mediates this splicing by maintaining the protein stability of splicing factor OsSR45, and both of them work together during abiotic stresses to regulate the RNA metabolism [120].
2.2.2. Circular RNAs
Circular RNAs (circRNAs), which are produced through back-splicing in pre-mRNA, play important roles in different plant biological processes and can regulate gene expression through modulating AS and sequestering miRNAs [121]. The functional role of these endogenous noncoding RNAs in rice stress responses is not fully explored. A genome-wide study has revealed the potential role of circRNAs in response to phosphate-starvation stress in rice [122]. The study found differential expression of 27 exoniccircRNAs under phosphate-deficient conditions, with six being significantly up-regulated and 21 down-regulated. Further, the expression of some of these circRNAs was correlated with that of their parent genes, implying their coregulation under phosphate-starvation stress. A recently developed comprehensive database for crop abiotic stress-responsive circRNAs hosts 63,048 rice circRNAs responsive to drought, salt, cold and other stresses, including their thorough details [123]. This database can be used to gain critical insights into the role of circRNAs in rice abiotic stress tolerance. Another database, called AsmiR (http://forestry.fafu.edu.cn/bioinfor/db/ASmiR, accessed on 20 February 2020), identifies the miRNA alternatively targets spliced transcripts and circRNAs in several plants, including rice [124].
Although circRNAs are regarded as noncoding, recent studies on mammals and plants provide evidence for certain protein-coding circRNAs [125][126].
2.2.3. AS and Splicing Factors
AS is precisely regulated by a distinct group of RNA binding proteins, known as trans-splicing factors, which bind to specific RNA sequence signals called cis-splicing elements. Among these factors are SR proteins, which play a crucial role in the assembly and recruitment of spliceosome to splice sites by interacting with splicing elements and other splicing factors [20][21]. Plants have a higher number of SR genes than animals [127], perhaps as a way of building the capacity to respond to wide-ranging unavoidable environmental cues. SR proteins are closely involved in rice abiotic stress responses. For example, analysis of rice sr mutants has revealed that SR splicing factors have a regulatory role in maintaining the nutrient homeostasis in rice; with RS29 for Mn-, RS33 for Zn-, and many SR proteins (e.g., SR40, SCL57, SCL25) being essential for P-homeostasis in rice shoots [11]. In particular, the P remobilization from old to young leaves is significantly reduced in sr40 and scl57 mutants, suggesting that these two proteins could be involved in phloem transport of phosphate by positively regulating the expression of phosphate mobilizing transporters such as PHT. Another splicing factor, CWC25, is essential for spliceosome activity by modulating its structure at the first transesterification reaction [119]. The functional analysis of CWC25 has revealed another example of the importance of AS in rice abiotic stress tolerance. The cwc25-knockout mutants exhibit reduced seed germination and shorter seedling roots as compared to wild-type under ABA and salt stress [119]. The spliceosome component, Ski-interacting protein (OsSKIPa), regulates cell viability and growth of rice under ABA, drought, and salt stress [117]. OsSKIPa-transgenic rice plants exhibit improved drought tolerance at seedling and reproductive stages, due to the high potential of scavenging the ROS and up-regulation of crucial stress-responsive genes, such as SNAC1, CBF2, PP2C, and RD22. OsSKIPa complements the lethal phenotype of yeast PRP45 (a splicing factor) mutant, indicating that OsSKIPa also functions as a splicing factor [117]. Functional analysis of yet another spliceosome-associated protein, DEAD-box RNA Helicase42 (OsRH42), has revealed its role in regulating effective pre-mRNA splicing under cold stress. In the cold-stressed OsRH42-knockdown and -overexpression plants, genome-wide AS, as well as AS of a subset of cold-responsive genes (e.g., ALKALINE INVERTASE6 and DST COACTIVATOR1) is severely affected, resulting in retarded growth and cold-sensitivity [128]. The availability of single and multiple mutants of rice SR genes that were generated using the CRISPR/Cas9 technology should expedite the functional analyses of rice SR proteins in biotic and abiotic stress responses [129].
SR proteins being the master-regulators of AS, implies that they could themselves be tightly regulated. In fact, the SR transcripts undergo AS under ABA, NaCl, heat, and cold stresses, with their alternative transcripts exhibiting differential expression in three different rice ecotypes under these stresses [118]. Besides, most of their isoforms harbor PTC leading to the truncated products, which might act as an extra layer of regulation for the expression of their target genes under stress.
Collectively, these examples illustrate that splicing factors and spliceosome-associated components can be controlled by various abiotic stresses and can, in turn, regulate the rice responses to these stresses.
References
- Bhandari, H.; Mishra, A.K. Impact of demographic transformation on future rice farming in Asia. Outlook Agric. 2018, 47, 125–132.
- FAO. Crop Prospects and Food Situation-Quarterly Global Report No. 1; FAO: Rome, Italy, 2021.
- Liu, W.; Wang, G.-L. Plant innate immunity in rice: A defense against pathogen infection. Natl. Sci. Rev. 2016, 3, 295–308.
- Ganie, S.A.; Ahammed, G.J.; Wani, S.H. Vascular plant one zinc-finger (VOZ) transcription factors: Novel regulators of abiotic stress tolerance in rice (Oryza sativa L.). Genet. Resour. Crop. Evol. 2020, 67, 799–807.
- Motion, G.B.; Amaro, T.M.M.M.; Kulagina, N.; Huitema, E. Nuclear processes associated with plant immunity and pathogen susceptibility. Brief. Funct. Genom. 2015, 14, 243–252.
- Haak, D.C.; Fukao, T.; Grene, R.; Hua, Z.; Ivanov, R.; Perrella, G.; Li, S. Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 2017, 8, 1564.
- Guerra, D.; Crosatti, C.; Khoshro, H.H.; Mastrangelo, A.M.; Mica, E.; Mazzucotelli, E. Post-transcriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms. Front. Plant Sci. 2015, 6, 57.
- Ganie, S.A. RNA chaperones: Potential candidates for engineering salt tolerance in rice. Crop. Sci. 2020, 60, 530–540.
- Laloum, T.; Martín, G.; Duque, P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 2018, 23, 140–150.
- Rigo, R.; Bazin, J.; Crespi, M.; Charon, C. Alternative splicing in the regulation of plant–microbe interactions. Plant Cell Physiol. 2019, 60, 1906–1916.
- Dong, C.; He, F.; Berkowitz, O.; Liu, J.; Cao, P.; Tang, M.; Shi, H.; Wang, W.; Li, Q.; Shen, Z.; et al. Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 2018, 30, 2267–2285.
- Jabre, I.; Reddy, A.S.; Kalyna, M.; Chaudhary, S.; Khokhar, W.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Does co-transcriptional regulation of alternative splicing mediate plant stress responses? Nucleic Acids Res. 2019, 47, 2716–2726.
- Chaudhary, S.; Jabre, I.; Reddy, A.S.; Staiger, D.; Syed, N.H. Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 2019, 24, 496–506.
- Zalabák, D.; Ikeda, Y. First Come, First Served: Sui Generis Features of the First Intron. Plants 2020, 9, 911.
- Breitbart, R.E.; Andreadis, A.; Nadal-Ginard, B. Alternative splicing: A ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem. 1987, 56, 467–495.
- Reddy, A.S.N. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294.
- Ner-Gaon, H.; Halachmi, R.; Savaldi-Goldstein, S.; Rubin, E.; Ophir, R.; Fluhr, R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004, 39, 877–885.
- Reddy, A.S.; Rogers, M.F.; Richardson, D.N.; Hamilton, M.; Ben-Hur, A. Deciphering the plant splicing code: Experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front. Plant Sci. 2012, 3, 18.
- Palusa, S.G.; Reddy, A.S.N. Differential recruitment of splice variants from SR pre-mRNAs to polysomes during development and in response to stresses. Plant Cell Physiol. 2015, 56, 421–427.
- Reddy, A.S.N.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683.
- Staiger, D.; Brown, J.W.S. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656.
- Black, D.L. Mechanisms of alternative pre-messenger rna splicing. Annu. Rev. Biochem. 2003, 72, 291–336.
- Barta, A.; Kalyna, M.; Reddy, A.S.N. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 2010, 22, 2926–2929.
- Reddy, A.S.N.; Huang, J.; Syed, N.H.; Ben-Hur, A.; Dong, S.; Gu, L. Decoding co-/post-transcriptional complexities of plant transcriptomes and epitranscriptome using next-generation sequencing technologies. Biochem. Soc. Trans. 2020, 48, 2399–2414.
- Ullah, F.; Hamilton, M.; Reddy, A.S.N.; Ben-Hur, A. Exploring the relationship between intron retention and chromatin accessibility in plants. BMC Genom. 2018, 19.
- Wang, X.; Hu, L.; Wang, X.; Li, N.; Xu, C.; Gong, L.; Liu, B. DNA methylation affects gene alternative splicing in plants: An example from rice. Mol. Plant 2016, 9, 305–307.
- Pajoro, A.; Severing, E.; Angenent, G.C.; Immink, R.G. Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol. 2017, 18.
- Yu, X.; Meng, X.; Liu, Y.; Wang, X.; Wang, T.-J.; Zhang, A.; Li, N.; Qi, X.; Liu, B.; Xu, Z.-Y. The chromatin remodeler ZmCHB101 impacts alternative splicing contexts in response to osmotic stress. Plant Cell Rep. 2018, 38, 131–145.
- Yan, K.; Liu, P.; Wu, C.-A.; Yang, G.-D.; Xu, R.; Guo, Q.-H.; Huang, J.-G.; Zheng, C.-C. Stress-Induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 2012, 48, 521–531.
- Campo, S.; Peris-Peris, C.; Siré, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; San Segundo, B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227.
- Jia, F.; Rock, C.D. MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol. Biol. 2013, 81, 447–460.
- Meng, Y.; Shao, C.; Ma, X.; Wang, H. Introns targeted by plant microRNAs: A possible novel mechanism of gene regulation. Rice 2013, 6.
- Park, S.-Y.; Grabau, E. Bypassing miRNA-mediated gene regulation under drought stress: Alternative splicing affects CSD1 gene expression. Plant Mol. Biol. 2017, 95, 243–252.
- Zhang, Y.; Rahmani, R.S.; Yang, X.; Chen, J.; Shi, T. Integrative expression network analysis of microRNA and gene isoforms in sacred lotus. BMC Genom. 2020, 21, 1–13.
- Chen, L.; Tovar-Corona, J.M.; Urrutia, A.O. Alternative Splicing: A Potential Source of Functional Innovation in the Eukaryotic Genome. Int. J. Evol. Biol. 2012, 2012, 1–10.
- Brogna, S.; McLeod, T.; Petric, M. The meaning of NMD: Translate or perish. Trends Genet. 2016, 32, 395–407.
- Ohtani, M.; Wachter, A. NMD-based gene regulation—A strategy for fitness enhancement in plants? Plant Cell Physiol. 2019, 60, 1953–1960.
- Gracz, J. Alternative splicing in plant stress response. BioTechnologia 2016, 1, 9–17.
- Liu, J.; Sun, N.; Liu, M.; Liu, J.; Du, B.; Wang, X.; Qi, X. An autoregulatory loop controlling Arabidopsis HsfA2 expression: Role of heat shock-induced alternative splicing. Plant Physiol. 2013, 162, 512–521.
- Mastrangelo, A.M.; Marone, D.; Laidò, G.; De Leonardis, A.M.; De Vita, P. Alternative splicing: Enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012, 185–186, 40–49.
- Srivastava, A.K.; Lu, Y.; Zinta, G.; Lang, Z.; Zhu, J.-K. UTR-dependent control of gene expression in plants. Trends Plant Sci. 2018, 23, 248–259.
- Punzo, P.; Grillo, S.; Batelli, G. Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 2020, 48, 2117–2126.
- Palusa, S.G.; Ali, G.S.; Reddy, A.S.N. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107.
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative Splicing and Protein Diversity: Plants Versus Animals. Front. Plant Sci. 2019, 10.
- Chen, M.-X.; Zhang, K.-L.; Zhang, M.; Das, D.; Fang, Y.-M.; Dai, L.; Zhang, J.; Zhu, F.-Y. Alternative splicing and its regulatory role in woody plants. Tree Physiol. 2020, 40, 1475–1486.
- Ling, Y.; Serrano, N.; Gao, G.; Atia, M.; Mokhtar, M.; Woo, Y.H.; Bazin, J.; Veluchamy, A.; Benhamed, M.; Crespi, M.; et al. Thermopriming triggers Splicing memory in Arabidopsis. J. Exp. Bot. 2018, 69, 2659–2675.
- Martin, L.B.; Fei, Z.; Giovannoni, J.J.; Rose, J.K. Catalyzing plant science research with RNA-seq. Front. Plant Sci. 2013, 4.
- Jain, M. Next-generation sequencing technologies for gene expression profiling in plants. Brief. Funct. Genom. 2011, 11, 63–70.
- Takahagi, K.; Uehara-Yamaguchi, Y.; Yoshida, T.; Sakurai, T.; Shinozaki, K.; Mochida, K.; Saisho, D. Analysis of single nucleotide polymorphisms based on RNA sequencing data of diverse bio-geographical accessions in barley. Sci. Rep. 2016, 6, 1–11.
- He, F.; Liu, Q.; Zheng, L.; Cui, Y.; Shen, Z.; Zheng, L. RNA-seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress. Front. Plant Sci. 2015, 6.
- Wei, H.; Lou, Q.; Xu, K.; Yan, M.; Xia, H.; Ma, X.; Yu, X.; Luo, L. Alternative splicing complexity contributes to genetic improvement of drought resistance in the rice maintainer HuHan2B. Sci. Rep. 2017, 7, 1–13.
- Li, N.; Liu, H.; Sun, J.; Zheng, H.; Wang, J.; Yang, L.; Zhao, H.; Zou, D. Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci. Rep. 2018, 8, 1–16.
- Phule, A.S.; Barbadikar, K.M.; Maganti, S.M.; Seguttuvel, P.; Subrahmanyam, D.; Babu, M.B.; Kumar, P.A. RNA-seq reveals the involvement of key genes for aerobic adaptation in rice. Sci. Rep. 2019, 9, 1–10.
- Razzaque, S.; Elias, S.M.; Haque, T.; Biswas, S.; Jewel, G.M.; Rahman, S.; Weng, X.; Ismail, A.M.; Walia, H.; Juenger, T.E.; et al. Gene expression analysis associated with salt stress in a reciprocally crossed rice population. Sci. Rep. 2019, 9, 1–17.
- Liu, G.; Zha, Z.; Cai, H.; Qin, D.; Jia, H.; Liu, C.; Qiu, D.; Zhang, Z.; Wan, Z.; Yang, Y.; et al. Dynamic transcriptome analysis of anther response to heat stress during anthesis in thermotolerant rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1155.
- Zhang, Z.; Xiao, B. Comparative alternative splicing analysis of two contrasting rice cultivars under drought stress and association of differential splicing genes with drought response QTLs. Euphytica 2018, 214.
- Mansuri, R.M.; Shobbar, Z.S.; Jelodar, N.B.; Ghaffari, M.R.; Nematzadeh, G.A.; Asari, S. Dissecting molecular mechanisms underlying salt tolerance in rice: A comparative transcriptional profiling of the contrasting genotypes. Rice 2019, 12.
- Fu, L.; Shen, Q.; Kuang, L.; Wu, D.; Zhang, G. Transcriptomic and alternative splicing analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Environ. Exp. Bot. 2019, 166, 103810.
- Shankar, R.; Bhattacharjee, A.; Jain, M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 2016, 6, 1–15.
- Junior, A.T.D.; Farias, D.D.; dos Santos, R.S.; do Amaral, M.N.; Arge, L.W.P.; Oliveira, D.D.C.; de Oliveira, A.C. The quest for more tolerant rice: How high concentrations of iron affect alternative splicing? Transcr. Open Access 2015, 3.
- Chen, M.-X.; Zhu, F.-Y.; Wang, F.-Z.; Ye, N.-H.; Gao, B.; Chen, X.; Zhao, S.-S.; Fan, T.; Cao, Y.-Y.; Liu, T.-Y.; et al. Alternative splicing and translation play important roles in hypoxic germination in rice. J. Exp. Bot. 2019, 70, 817–833.
- Sampangi-Ramaiah, M.H.; Ravishankar, K.V.; Nataraja, K.N.; Uma Shaanker, R. Endophytic fungus, Fusarium sp. reduces alternative splicing events in rice plants under salinity stress. Plant Physiol. Rep. 2019, 24, 487–495.
- Li, Y.-F.; Zheng, Y.; Vemireddy, L.R.; Panda, S.K.; Jose, S.; Ranjan, A.; Panda, P.; Govindan, G.; Cui, J.; Wei, K.; et al. Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. BMC Genom. 2018, 19.
- Ganie, S.A.; Ahammed, G.J. Dynamics of cell wall structure and related genomic resources for drought tolerance in rice. Plant Cell Rep. 2021, 40, 437–459.
- Ganie, S.A.; Wani, S.H.; Henry, R.; Hensel, G. Improving rice salt tolerance by precision breeding in a new era. Curr. Opin. Plant Biol. 2021, 60, 101996.
- Zhang, T.; Huang, L.; Wang, Y.; Wang, W.; Zhao, X.; Zhang, S.; Zhang, J.; Hu, F.; Fu, B.; Li, Z. Differential transcriptome profiling of chilling stress response between shoots and rhizomes of Oryza longistaminata using RNA sequencing. PLoS ONE 2017, 12.
- Deng, Q.; Bai, L.; Dai, L.; Chen, Y.; Fang, J.; Xie, J.; Luo, X. Identification of phosphorus stress related proteins in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff.) using label-free quantitative proteomic analysis. Res. Square 2020.
- Wei, H.; Lou, Q.; Xu, K.; Zhou, L.; Chen, S.; Chen, L.; Luo, L. Pattern of alternative splicing different associated with difference in rooting depth in rice. Plant Soil 2020, 449, 233–248.
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870.
- Matsukura, S.; Mizoi, J.; Yoshida, T.; Todaka, D.; Ito, Y.; Maruyama, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom. 2010, 283, 185–196.
- Magaraggia, F.; Solinas, G.; Valle, G.; Giovinazzo, G.; Coraggio, I. Maturation and translation mechanisms involved in the expression of a myb gene of rice. Plant Mol. Biol. 1997, 35, 1003–1008.
- Cheng, Q.; Zhou, Y.; Liu, Z.; Zhang, L.; Song, G.; Guo, Z.; Wang, W.; Qu, X.; Zhu, Y.; Yang, D. An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. 2015, 17, 419–429.
- Cotsaftis, O.; Plett, D.; Shirley, N.; Tester, M.; Hrmova, M. A Two-Staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 2012, 7, e39865.
- Kong, J.; Gong, J.-M.; Zhang, Z.-G.; Zhang, J.-S.; Chen, S.-Y. A new AOX homologous gene OsIM1 from rice (Oryza sativa L.) with an alternative splicing mechanism under salt stress. Theor. Appl. Genet. 2003, 107, 326–331.
- Cordeiro, A.M.; Figueiredo, D.D.; Tepperman, J.; Borba, A.R.; Lourenço, T.; Abreu, I.A.; Ouwerkerk, P.B.F.; Quail, P.H.; Margarida Oliveira, M.; Saibo, N.J.M. Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 393–404.
- Xu, H.; Li, X.; Zhang, H.; Wang, L.; Zhu, Z.; Gao, J.; Li, C.; Zhu, Y. High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell Environ. 2020, 43, 1879–1896.
- Mimida, N.; Kitamoto, H.; Osakabe, K.; Nakashima, M.; Ito, Y.; Heyer, W.-D.; Toki, S.; Ichikawa, H. Two Alternatively Spliced Transcripts generated From OSMUS81, a Rice homolog of Yeast MUS81, are Up-Regulated by DNA-Damaging Treatments. Plant Cell Physiol. 2007, 48, 648–654.
- Lee, A.; Lee, S.; Jung, W.; Park, H.; Lim, B.; Kim, H.-S.; Ahn, J.; Cho, H. The OsCYP19-4 Gene Is Expressed as Multiple Alternatively Spliced Transcripts Encoding Isoforms with Distinct Cellular Localizations and PPIase Activities under Cold Stress. Int. J. Mol. Sci. 2016, 17, 1154.
- Li, R.; Wang, W.; Li, F.; Wang, Q.; Wang, S.; Xu, Y.; Chen, F. Response of alternative splice isoforms of OsRad9 gene from Oryza sativa to environmental stress. Z. Naturforsch. C 2017, 72, 325–334.
- Sripinyowanich, S.; Chamnanmanoontham, N.; Udomchalothorn, T.; Maneeprasopsuk, S.; Santawee, P.; Buaboocha, T.; Qu, L.-J.; Gu, H.; Chadchawan, S. Overexpression of a Partial fragment of the salt-responsive gene OsNUC1 enhances salt adaptation in transgenic Arabidopsis thaliana and rice (Oryza sativa L.) during salt stress. Plant Sci. 2013, 213, 67–78.
- Koo, S.C.; Yoon, H.W.; Kim, C.Y.; Moon, B.C.; Cheong, Y.H.; Han, H.J.; Lee, S.M.; Kang, K.Y.; Kim, M.C.; Lee, S.Y.; et al. Alternative splicing of the OsBWMK1 gene generates three transcript variants showing differential subcellular localizations. Biochem. Biophys. Res. Commun. 2007, 360, 188–193.
- Sakuraba, Y.; Kim, D.; Han, S.-H.; Kim, S.-H.; Piao, W.; Yanagisawa, S.; An, G.; Paek, N.-C. Multilayered regulation of membrane-bound ONAC054 is essential for abscisic acid-induced leaf senescence in rice. Plant Cell 2020, 32, 630–649.
- Lee, C.-Y.; Tsai, Y.-C. Light-Regulated alternative splicing of Pseudo-histidine Phosphotransfer Protein 3 in Oryza sativa. J. Plant Growth Regul. 2019, 38, 1215–1227.
- Yang, S.-Y.; Lu, W.-C.; Ko, S.-S.; Sun, C.-M.; Hung, J.-C.; Chiou, T.-J. Upstream open reading frame and phosphate-regulated expression of rice OsNLA1 controls phosphate transport and reproduction. Plant Physiol. 2020, 182, 393–407.
- Zhang, Y.-M.; Yan, Y.-S.; Wang, L.-N.; Yang, K.; Xiao, N.; Liu, Y.-F.; Fu, Y.-P.; Sun, Z.-X.; Fang, R.-X.; Chen, X.-Y. A novel rice gene, NRR responds to macronutrient deficiency and regulates root growth. Mol. Plant 2012, 5, 63–72.
- Larkin, P.D.; Park, W.D. Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol. Biol. 1999, 40, 719–727.
- Amin, U.S.; Biswas, S.; Elias, S.M.; Razzaque, S.; Haque, T.; Malo, R.; Seraj, Z.I. Enhanced salt tolerance conferred by the complete 2.3 kb cDNA of the rice vacuolar Na+/H+ antiporter gene compared to 1.9 kb coding region with 5′ UTR in transgenic lines of rice. Front. Plant Sci. 2016, 7.
- Costanzo, S.; Jia, Y. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci. 2009, 177, 468–478.
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481.
- Wang, J.; Tian, D.; Gu, K.; Yang, X.; Wang, L.; Zeng, X.; Yin, Z. Induction of Xa10-like genes in rice cultivar Nipponbare confers disease resistance to rice bacterial blight. Mol. Plant Microbe Interact. 2017, 30, 466–477.
- Liu, J.; Chen, X.; Liang, X.; Zhou, X.; Yang, F.; Liu, J.; He, S.Y.; Guo, Z. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in Pathogen defense. Plant Physiol. 2016, 171, 1427–1442.
- Duan, L.; Xiao, W.; Xia, F.; Liu, H.; Xiao, J.; Li, X.; Wang, S. Two different transcripts of A LAMMER Kinase Gene play Opposite roles in disease resistance. Plant Physiol. 2016, 172, 1959–1972.
- Liu, D.; Shi, S.; Hao, Z.; Xiong, W.; Luo, M. OsbZIP81, A Homologue of Arabidopsis VIP1, May Positively Regulate JA Levels by Directly Targetting the Genes in JA Signaling and Metabolism Pathway in Rice. Int. J. Mol. Sci. 2019, 20, 2360.
- Hu, X.; Zhang, H.; Li, G.; Yang, Y.; Zheng, Z.; Song, F. Ectopic expression of a rice protein phosphatase 2C gene OsBIPP2C2 in tobacco improves disease resistance. Plant Cell Rep. 2009, 28, 985–995.
- Zhang, X.-B.; Feng, B.-H.; Wang, H.-M.; Xu, X.; Shi, Y.-F.; He, Y.; Chen, Z.; Sathe, A.P.; Shi, L.; Wu, J.-L. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 2018, 60, 160–172.
- Feng, W.; Hongbin, W.; Bing, L.; Jinfa, W. Cloning and characterization of a novel splicing isoform of the iron-superoxide dismutase gene in rice (Oryza sativa L.). Plant Cell Rep. 2006, 24, 734–742.
- Stracke, R.; Favory, J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010, 33, 88–103.
- Zhao, B.; Tang, Y.; Zhang, B.; Wu, P.; Li, M.; Xu, X.; Wu, G.; Jiang, H.; Chen, Y. The Temperature-Dependent Retention of Introns in GPI8 Transcripts Contributes to a Drooping and Fragile Shoot Phenotype in Rice. Int. J. Mol. Sci. 2020, 21, 299.
- Das, N.; Bhattacharya, S.; Bhattacharyya, S.; Maiti, M.K. Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2 involved in mitigation of cadmium and arsenic stresses. Plant Mol. Biol. 2017, 94, 167–183.
- Sellamuthu, G.; Jegadeeson, V.; Sajeevan, R.S.; Rajakani, R.; Parthasarathy, P.; Raju, K.; Shabala, L.; Chen, Z.-H.; Zhou, M.; Sowdhamini, R.; et al. Distinct evolutionary origins of intron retention splicing events in NHX1 antiporter transcripts relate to sequence specific distinctions in Oryza species. Front. Plant Sci. 2020, 11.
- Koo, S.C.; Choi, M.S.; Chun, H.J.; Park, H.C.; Kang, C.H.; Shim, S.I.; Chung, J.I.; Cheong, Y.H.; Lee, S.Y.; Yun, D.-J.; et al. Identification and characterization of alternative promoters of the rice MAP kinase gene OsBWMK1. Mol. Cells 2009, 27, 467–473.
- Kumar, S.; Asif, M.H.; Chakrabarty, D.; Tripathi, R.D.; Dubey, R.S.; Trivedi, P.K. Comprehensive analysis of regulatory elements of the promoters of rice sulfate transporter gene family and functional characterization of OsSul1;1 promoter under different metal stress. Plant Signal. Behav. 2015, 10.
- Khurana, N.; Chauhan, H.; Khurana, P. Expression analysis of A Heat-inducible, Myo-inositol-1-phosphate synthase (MIPS) gene from wheat and the alternatively spliced variants of rice and Arabidopsis. Plant Cell Rep. 2011, 31, 237–251.
- Kumar, S.; Asif, M.H.; Chakrabarty, D.; Tripathi, R.D.; Trivedi, P.K. Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funct. Integr. Genom. 2011, 11, 259–273.
- Wang, H.; Bian, M.; Yang, Z.; Lin, C.; Shi, W. Preliminary functional analysis of the Isoforms of OsHsfA2a (Oryza sativa L.) generated by alternative splicing. Plant Mol. Biol. Rep. 2013, 31, 38–46.
- Fang, Z. Differential expression pattern of splice variants of amino acid transporter genes from rice grown under various nitrogen conditions and during development. Int. J. Agric. Biol. 2017, 19, 1246–1258.
- Gupta, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front. Plant Sci. 2017, 8.
- Almadanim, M.C.; Gonçalves, N.M.; Rosa, M.T.G.; Alexandre, B.M.; Cordeiro, A.M.; Rodrigues, M.; Saibo, N.J.M.; Soares, C.M.; Romão, C.V.; Oliveira, M.M.; et al. The rice cold-responsive calcium-dependent protein kinase OsCPK17 is regulated by alternative splicing and post-translational modifications. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 231–246.
- Xiong, L.; Yang, Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid–inducible mitogen-activated protein kinase. Plant Cell 2003, 15, 745–759.
- Zeng, C.; Hamada, M. RNA-Seq analysis reveals localization-associated alternative splicing across 13 cell lines. Genes 2020, 11, 820.
- Lu, S.-J.; Yang, Z.-T.; Sun, L.; Sun, L.; Song, Z.-T.; Liu, J.-X. Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol. Plant 2012, 5, 504–514.
- Kornblihtt, A.R. Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 2005, 17, 262–268.
- Lin, W.-Y.; Huang, T.-K.; Chiou, T.-J. NITROGEN limitation adaptation, a target of MicroRNA827, Mediates degradation of Plasma Membrane–Localized Phosphate transporters to Maintain Phosphate homeostasis in Arabidopsis. Plant Cell 2013, 25, 4061–4074.
- Tanaka, M.; Sotta, N.; Yamazumi, Y.; Yamashita, Y.; Miwa, K.; Murota, K.; Chiba, Y.; Hirai, M.Y.; Akiyama, T.; Onouchi, H.; et al. The Minimum Open Reading Frame, AUG-Stop, Induces Boron-Dependent Ribosome Stalling and mRNA Degradation. Plant Cell 2016, 28, 2830–2849.
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem. Biophys. Res. Commun. 2007, 360, 307–313.
- Rosa, M.T.; Almeida, D.M.; Pires, I.S.; da Rosa Farias, D.; Martins, A.G.; da Maia, L.C.; de Oliveira, A.C.; Saibo, N.J.; Oliveira, M.M.; Abreu, I.A. Insights into the transcriptional And Post-transcriptional regulation of the rice SUMOylation machinery and into the role of two rice SUMO proteases. BMC Plant Biol. 2018, 18.
- Hou, X.; Xie, K.; Yao, J.; Qi, Z.; Xiong, L. A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc. Natl. Acad. Sci. USA 2009, 106, 6410–6415.
- Zhang, P.; Deng, H.; Xiao, F.; Liu, Y. Alterations of Alternative Splicing Patterns of Ser/Arg-Rich (SR) Genes in Response to Hormones and Stresses Treatments in Different Ecotypes of Rice (Oryza sativa). J. Integr. Agric. 2013, 12, 737–748.
- Kababji, A.M. Targeted Mutagenesis and Functional Analysis of CWC25 Splicing Factor in Rice via CRISPR/Cas9. Ph.D. Thesis, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia, 2019.
- Park, H.J.; You, Y.N.; Lee, A.; Jung, H.; Jo, S.H.; Oh, N.; Kim, H.S.; Lee, H.J.; Kim, J.K.; Kim, Y.S.; et al. OsFKBP20-1b interacts with the splicing factor OsSR45 and participates in the environmental stress response at the post-transcriptional level in rice. Plant J. 2020, 102, 992–1007.
- Zhang, P.; Li, S.; Chen, M. Characterization and function of circular RNAs in plants. Front. Mol. Biosci. 2020, 7.
- Ye, C.Y.; Chen, L.; Liu, C.; Zhu, Q.H.; Fan, L. Widespread noncoding circular RNAs in plants. New Phytol. 2015, 208, 88–95.
- Wang, K.; Wang, C.; Guo, B.; Song, K.; Shi, C.; Jiang, X.; Wang, K.; Tan, Y.; Wang, L.; Wang, L.; et al. CropCircDB: A comprehensive circular RNA resource for crops in response to abiotic stress. Database 2019, 2019.
- Wang, H.; Wang, H.; Zhang, H.; Liu, S.; Wang, Y.; Gao, Y.; Xi, F.; Zhao, L.; Liu, B.; Reddy, A.S.; et al. The interplay between microRNA and alternative splicing of linear and circular RNAs in eleven plant species. Bioinformatics 2019, 35, 3119–3126.
- Schneider, T.; Bindereif, A. Circular RNAs: Coding or noncoding? Cell Res. 2017, 27, 724–725.
- Wang, Y.; Wang, H.; Xi, F.; Wang, H.; Han, X.; Wei, W.; Zhang, H.; Zhang, Q.; Zheng, Y.; Zhu, Q.; et al. Profiling of circular RNA N6 -methyladenosine in moso bamboo (Phyllostachys edulis) using nanopore-based direct RNA sequencing. J. Integr. Plant Biol. 2020, 62, 1823–1838.
- Richardson, D.N.; Rogers, M.F.; Labadorf, A.; Ben-Hur, A.; Guo, H.; Paterson, A.H.; Reddy, A.S. Comparative Analysis of Serine/Arginine-Rich Proteins across 27 Eukaryotes: Insights into Sub-Family Classification and Extent of Alternative Splicing. PLoS ONE 2011, 6.
- Lu, C.-A.; Huang, C.-K.; Huang, W.-S.; Huang, T.-S.; Liu, H.-Y.; Chen, Y.-F. DEAD-Box RNA Helicase 42 plays a critical role in pre-mRNA splicing under cold stress. Plant Physiol. 2020, 182, 255–271.
- Butt, H.; Piatek, A.; Li, L.; Reddy, A.S.N.; Mahfouz, M.M. Multiplex CRISPR mutagenesis of the Serine/arginine-rich (SR) gene family in rice. Genes 2019, 10, 596.




