- Subjects: Agriculture, Dairy & Animal Science
- |
- Contributors:
- Tohru Minamino ,
- Shuichi Nakamura
- bacterial flagellum
- chemotaxis
- ion motive force
- ion channel
- mechanochemical coupling
- molecular motor
- motility
- torque generation
This video is adapted from 10.3390/biom9070279
Bacterial motility is an extremely intriguing topic from various scientific aspects. For example, motility can be a crucial virulence attribute for pathogenic bacteria, such as Salmonella enterica (hereafter referred to Salmonella) and Helicobacter pylori [1][2]. Bacterial motility also plays a significant role in mutualistic symbioses [3][4]. Furthermore, motile bacteria are also a representative example for understanding the underlying physical principles that form the basis of energy conversion, force generation and mechanochemical coupling mechanisms [5]. Active motilities of bacteria are represented by movement in liquid (e.g., swimming motility in Escherichia coli and Salmonella) and on solid surfaces (e.g., flagella-driven swarming motility in Proteus mirabilis and Vibrio parahaemolyticus, gliding motility in Mycoplasma mobile, and twitching motility in Pseudomonas aeruginosa), and passive motility is typically actin-based locomotion (e.g., Listeria monocytogenes and Shigella spp.) [6]. Since bacterial motility varies among bacterial species, bacteria utilize their own motility system optimized for their habitats.
E. coli and Salmonella use flagella viewable from the cell exterior as a thin, long, helical filament (Figure 1a). On the other hand, the flagella of spirochetes reside within the periplasmic space, and so they are called periplasmic flagella [7]. Whether the bacterial flagella are exposed to the cell exterior or are hidden within the cell body, the flagellum is divided into three structural parts: the basal body as a rotary motor, the hook as a universal joint and the filament as a molecular screw in common (Figure 1b), and flagellar formation and function involves more than 60 genes [8][9][10].
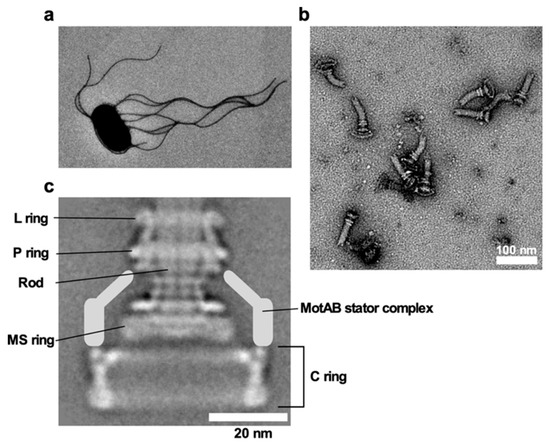
Figure 1. Salmonella flagellum. (a) Electron micrograph of Salmonella cell. The micrograph was taken at a magnification of ×1200. (b) Electron micrograph of hook-basal bodies isolated from Salmonella cells. (c) CryoEM image of purified basal body. Purified basal body consists of the L, P, MS and C rings and the rod. A dozen MotAB complex are associated with the basal body to act as a stator unit in the motor but is gone during purification.
The bacterial flagellar motor is powered by the transmembrane electrochemical gradient of ions, namely ion motive force (IMF) and rotates the flagellar filament to generate thrust to propel the cell body. The maximum motor speed reaches 300 revolutions per second in E. coli and Salmonella [11] and 1700 revolutions per second in a marine bacterium Vibrio alginolyticus [12]. Thus, the rotational speed of the flagellar motor is much faster than that of a manufactured car engine such as formula one car. The flagellar motor is composed of a rotor and multiple stator units. Each stator unit acts as a transmembrane ion channel to conduct cations such as protons (H+) or sodium ions (Na+) and applies force on the rotor [13][14].
The flagellar motors of E. coli and Salmonella rotate in both counterclockwise (CCW) and clockwise (CW) without changing the direction of ion flow. E. coli and Salmonella cells can swim in a straight line by bundling left-handed helical filaments behind the cell body (run) when all of them rotate in CCW direction. When one or multiple motors switch the direction of rotation from CCW to CW, the flagellar bundle is disrupted, enabling the cell to tumble and change the swimming direction. E. coli and Salmonella cells sense temporal changes in nutrients, environmental stimuli, and signaling molecules to coordinate the switching frequency of the motor. Transmembrane chemoreceptors, energy-related taxis sensors and intracellular phosphotransferase systems detect environmental signals and then convert them into intracellular signals. Then, an intracellular signal transduction system transmits the signals to the flagellar motor to switch the direction of motor rotation from CCW to CW. The cells repeat a run–tumble pattern to explore more favorable environments for their survival [15].
- Josenhans, C.; Suerbaum, S. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 2002, 291, 605–614.
- Lertsethtakarn, P.; Ottemann, K.M.; Hendrixson, D.R. Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 2011, 65, 389–410.
- Visick, K.L. An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol. Microbiol. 2009, 74, 782–789.
- Scharf, B.E.; Hynes, M.F.; Alexandre, G.M. Chemotaxis signaling systems in model beneficial plant–bacteria associations. Plant Mol. Biol. 2016, 90, 549–559.
- Ishikawa, T.; Yoshida, N.; Ueno, H.; Wiedeman, M.; Imai, Y.; Yamaguchi, T. Energy transport in a concentrated suspension of bacteria. Phys. Rev. Lett. 2011, 107, 028102.
- Jarrell, K.F.; McBride, M.J. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 2008, 6, 466–476.
- Charon, N.W.; Goldstein, S.F. Genetics of motility and chemotaxis of a fascinating group of bacteria: The spirochetes. Annu. Rev. Genet. 2002, 36, 47–73.
- Minamino, T.; Namba, K. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 2004, 7, 5–17.
- Terashima, H.; Kawamoto, A.; Morimoto, Y.V.; Imada, K.; Minamino, T. Structural differences in the bacterial flagellar motor among bacterial species. Biophys. Physicobiology 2017, 14, 191–198.
- Minamino, T.; Imada, K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015, 23, 267–274.
- Chen, X.; Berg, H.C. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys. J. 2000, 78, 1036–1041.
- Magariyama, Y.; Sugiyama, S.; Muramoto, K.; Maekawa, Y.; Kawagishi, I.; Imae, Y.; Kudo, S. Very fast flagellar rotation. Nature 1994, 371, 752.
- Morimoto, Y.V.; Minamino, T. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 2014, 4, 217–234.
- Minamino, T.; Terahara, N.; Kojima, S.; Namba, K. Autonomous control mechanism of stator assembly in the bacterial flagellar motor in response to changes in the environment. Mol. Microbiol. 2018, 109, 723–734.
- Berg, H.C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003, 72, 19–54.
