Selection for wheat (Triticum aestivum L.) grain quality is often costly and time-consuming
since it requires extensive phenotyping in the last phases of development of new lines and cultivars.
The development of high-throughput genotyping in the last decade enabled reliable and rapid
predictions of breeding values based only on marker information. Genomic selection (GS) is a method
that enables the prediction of breeding values of individuals by simultaneously incorporating all
available marker information into a model. The success of GS depends on the obtained prediction
accuracy, which is influenced by various molecular, genetic, and phenotypic factors, as well as the
factors of the selected statistical model.
- wheat quality
- genomic selection
- GEBV
- prediction accuracy
1. Introduction
The extensive development of high-throughput genotyping in the last couple of decades has led to the increasing use of molecular markers in plant breeding, which enabled the development of prediction methods based only on marker information such as genomic selection (GS) [1][2]. The first GS studies in wheat were published more than a decade ago [3][4]. The results of these studies showed that models based on genomic markers outperform models based only on pedigree relationships and that GS could successfully enhance rates of genetic gain, which provided a strong foundation for further research on GS in wheat. Later studies also showed that, if the traits of interest are complex and influenced by many quantitative trait loci (QTLs) each controlling a small proportion of phenotypic variation, GS will be more relevant than marker-assisted selection (MAS) [5][6].
Genomic selection is one of the newly developed methods that enables the prediction of breeding values of individuals by simultaneously incorporating all available marker information into a model [2]. Unlike other molecular breeding methods, GS does not require the identification of markers associated with QTLs (Quantitative Trait Loci) of traits of interest. GS attempts to capture total additive genetic variance based on the sum of the effects of a large number of genetic markers, encompassing all QTLs that contribute to trait variability [14]. Therefore, the underlying genetic control in GS is not necessarily known. In GS, training population (TP) is genotyped using one of the methods of high-throughput genotyping and phenotyped for desired traits in a target set of environments. Obtained data are used to train a model that will be applied to the breeding population (BP) of unphenotyped individuals (selection candidates) to calculate their genomic-estimated breeding values (GEBVs) using only the marker scores [2]. The most important advantage of GS over phenotypic selection (PS) is the increase of genetic gain due to the shortening of the selection cycle in breeding process by reducing the need for phenotyping [15][16].
Currently, the majority of researchers of GS in wheat consider grain yield and disease resistance as key traits for successful wheat production [7][8][9][10][11]. Such a strong focus on grain yield is understandable from a point of view where grain yield is not improving fast enough to fill the gap between production and projected demands in the near future [12]. Considering wheat’s role as the main ingredient in many different products fundamental to the nutrition of humankind, equal emphasis should be given to the quality traits, especially those related to the end-use quality of wheat products. An overview of GS research studies for traits such as grain yield, Fusarium head blight, stripe and brown rust resistance, plant height, days to heading, and preharvest sprouting (PHS) tolerance is given by Rutkoski et al. [13]. Despite their importance in the context of nutrition, research on GS for wheat quality traits is still scarce.
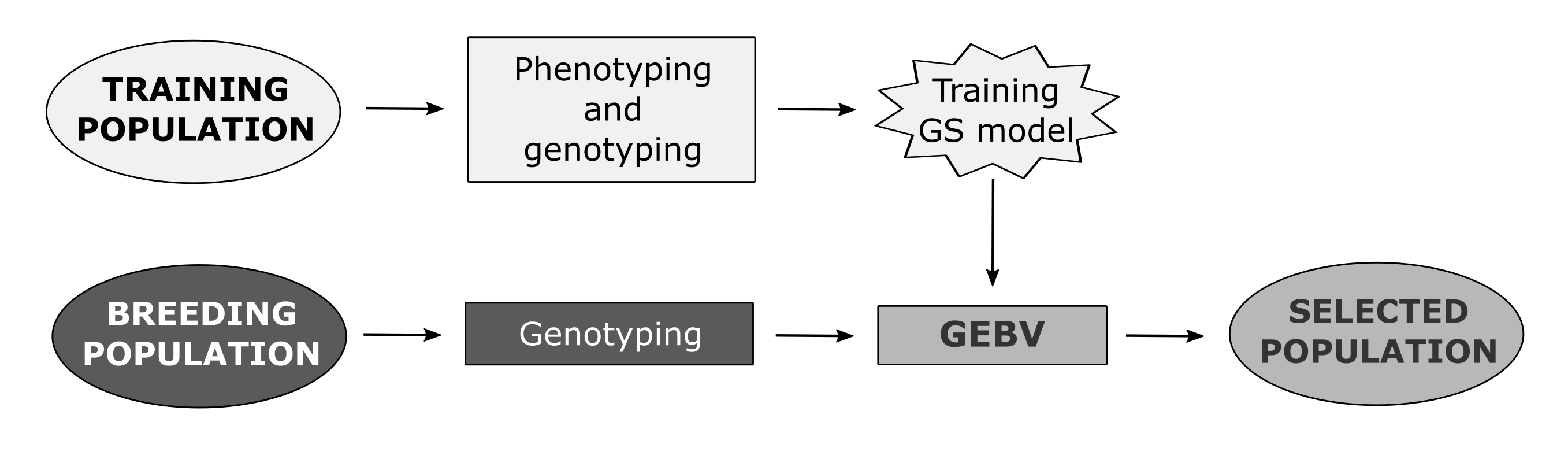
2. Factors Affecting Prediction Accuracies of Genomic Selection in Wheat
The prediction accuracy of GS is commonly estimated using cross-validation, in which a set of individuals that are both genotyped and phenotyped is divided into a training set (training population) and validation set (validation population, VP), with marker effects estimated in the training set used to predict GEBVs for the validation set [17]. The accuracy is then measured as the correlation between GEBVs and true breeding values (observed phenotypes) of individuals from the validation set. Prediction accuracy of GS is influenced by various molecular, genetic, and phenotypic factors, as well as the features of the selected statistical model. Three major factors that affect the GS accuracy are population structure, TP size, and marker density, the effects of which are highly interrelated [18][19]. Population structure can give rise to a false association between a marker and QTL, thus causing structure-generated LD, which can lead to overestimated genomic heritability and biased GS prediction accuracies [20][21]. As in other species, studies on wheat also showed that larger TP reduces bias and decreases the marker effect variance, thus resulting in higher prediction accuracy [22]. The interrelatedness of marker density and population structure seems to play an important role in optimizing GS in wheat. Namely, it has been shown that the higher the relatedness between TP and VP, the smaller the response to increased marker density [21].
3. Overview of Genomic Selection Research for Wheat Quality Improvement
The first GS study for wheat quality traits was published in 2011 [11]. The study was conducted in a population composed of multiple wheat families and showed that GS accuracy surpasses MAS accuracy for wheat quality traits by roughly 30% and that GS was about 95% as accurate as PS. Authors also concluded that, regardless of the inferiority when compared to PS, GS has the potential to increase genetic gain per unit of time and costs when applied in a breeding program. A study by Heffner et al. [23] based on two biparental wheat populations, examined the potential of GS to predict nine wheat quality traits. The authors of the study have found that the mean accuracy obtained by GS was 1.4 times greater than the one obtained by MAS, but that both GS and MAS were inferior to PS. However, those findings were expected due to the polygenic nature and medium to high heritability of all examined traits. Liu et al. [24] reported that, when predicting wheat hybrid performance for seven quality traits, GS extensively outperformed MAS, while giving similar results as PS in the case of higher relatedness of TP and VP. It was only in the case of lower relatedness of TP and VP that GS was preferred over PS, thus emphasizing the importance of additive effects in wheat quality traits. According to Battenfield et al. [25], genetic gain was 1.4 to 2.7 times higher when comparing GS to PS for processing and end-use quality traits since GS requires only marker data and a much larger population can be genotyped than phenotyped for wheat quality traits. Michel et al. [26] investigated the use of GS for predicting dough rheological traits in early generations and proved its substantial benefit over MAS. These findings imply that GS can capture more of the genetic variance of wheat quality traits when compared to MAS since it considers both small effect loci in addition to major QTLs. Nevertheless, all of the above-mentioned studies showed that the accuracy of GS for wheat quality traits is under the influence of many factors, with underlined heritability of the trait, genetic relationship between TP and VP, and size of the TP being the most important driving forces of GS accuracy.
As early as with the first studies of GS for wheat quality traits, it was demonstrated that TP size significantly impacts the GS accuracy. Overall, the size of the TP depends on the genetic relatedness between TP and VP. The more related the two populations are, the smaller the size of the TP will be needed to obtain satisfying GS prediction accuracies for wheat quality traits [27]. Battenfield et al. [25] also reported enhanced accuracy as a result of increasing TP size and random assignment of full-sibs to TP and VP, therefore, creating a greater genetic relationship. Considering that the phenotyping of wheat quality traits can be both costly and time-consuming, designing a TP that at the same time maximizes genetic diversity and enhances GS accuracy, while being small enough to achieve rapid phenotyping, is key for the successful implementation of GS in a breeding program [28].
Studies of GS for wheat quality traits investigating the effect of marker density all led to the same conclusion that the accuracy of the prediction enhances with increasing marker density until it reaches a plateau, after which a further increase in marker density has no effect on accuracy [23]. Since required marker density is primarily determined by the extent of LD in the examined population, it is assumed that lower marker density will be sufficient for closely related populations (e.g., biparental population) than for distant populations to achieve satisfying GS prediction accuracy. The interdependence of marker density and relatedness of TP and VP in the context of GS was illustrated in a study by Liu et al. [24] in which three scenarios representing low, intermediate, and high relatedness were used. In the case of lowly related TP and VP, the plateau was reached after ~3000 markers, whereas in the case of intermediate and highly related TP and VP, the plateau was reached at ~2000 and ~500 markers, respectively.
Numerous studies up to date showed that GS accuracy is strongly influenced by heritability, i.e., the fraction of the phenotypic variance of the trait due to genetic variance. Although there is no unambiguous categorization, the majority of studies on wheat categorize heritability values as low (<0.4), moderate (0.4–0.7), and high (>0.7) [11][23]. Generally, traits with high heritability show high GS accuracy and vice versa. The predictive ability of GS for wheat quality traits parallels their heritability which is often showed to be moderate to high. Studies on wheat quality traits showed that heritability was the main factor that affected the accuracy of GS [29]. Interestingly, not all highly heritable traits showed high GS accuracy [27]. Low heritability traits would require larger TP in order to attain the same prediction accuracy as in the case of traits with moderate to high heritability [30].
A broad range of models can be used to predict the phenotypic performance of wheat, but the performance of each model is interrelated with the genetic architecture of the examined trait and relatedness of TP and VP. The majority of GS studies for wheat quality traits used GBLUP (Genomic Best Linear Unbiased Predictor) and RRBLUP (Ridge-Regression Best Linear Unbiased Predictor) models, the performance of which was usually compared to one of the Bayesian models. Bayesian models usually require longer computation time compared to GBLUP or RRBLUP [2][31][32] but show no clear superiority over the other models across wheat quality traits [29][33], i.e., the accuracy of GS for wheat quality traits was generally not under the large influence of prediction model applied. Therefore, RRBLUP showed to be a model of choice in the majority of GS studies for wheat quality traits [27][31] due to its robustness and shorter computational time [34].
4. Multitrait Genomic Selection
Wheat quality traits can often be hard to improve, since they usually require a large amount of flour and/or labor to be invested, thus limiting the size of the TP that can be phenotyped which leads to insufficient GS accuracy. Incorporating additional phenotypic information in the multitrait approach for GS could help to overcome the problem of potentially low GS accuracy obtained for wheat quality traits. Multitrait GS data obtained utilizing rapid quality tests are used for predicting parameters of more laborious wheat quality tests. Rapid tests such as near-infrared (NIR) and nuclear magnetic resonance (NMR) methods are less labor-intensive and require a small amount of flour. It has been proved that incorporating NIR and NMR data into the multitrait approach increases the accuracy of GS for some wheat quality traits (accuracy ranged between 0 and 0.47, and between 0 and 0.69 in a single-trait approach and multitrait approach, respectively) [30]. Incorporating easily obtained gluten peak indices into multitrait GS analysis improved average prediction accuracy by roughly 20% in comparison to single-trait GS for dough rheology traits [35]. Lado et al. [36] showed that no multitrait model used performed better than a single-trait model, but that using highly correlated traits in multitrait GS for wheat quality allows reduction of TP up to 30% without significantly affecting the predictive ability of the model. Further research studies showed that using different GS indices in simultaneous selection for yield and wheat quality traits still does not outperform single-trait prediction, but suggested that simultaneous improvement of yield and wheat quality should target protein quality [37]. A significant gain of multitrait approach is expected only for low heritable traits that are incorporated with high heritable traits, between which high genetic correlation exists [38]. Data for traits incorporating together in a multitrait analysis must be already available or easy to obtain on a large number of samples in a short period of time [39].
This entry is adapted from the peer-reviewed paper 10.3390/plants10040745
References
- José Crossa; Yoseph Beyene; Semagn Kassa; Paulino Pérez; John M. Hickey; Charles Chen; Gustavo De Los Campos; Juan Burgueño; Vanessa S. Windhausen; Ed Buckler; et al.Jean-Luc JanninkMarco A. Lopez CruzRaman Babu Genomic Prediction in Maize Breeding Populations with Genotyping-by-Sequencing. G3 Genes|Genomes|Genetics 2013, 3, 1903-1926, 10.1534/g3.113.008227.
- T H Meuwissen; B J Hayes; M E Goddard; Prediction of total genetic value using genome-wide dense marker maps.. Genetics 2001, 157, 1819-1829.
- Gustavo De Los Campos; Hugo Naya; Daniel Gianola; José Crossa; Andrés Legarra; Eduardo Manfredi; Kent Weigel; José Miguel Cotes; Predicting Quantitative Traits With Regression Models for Dense Molecular Markers and Pedigree. Genetics 2009, 182, 375-385, 10.1534/genetics.109.101501.
- José Crossa; Gustavo De Los Campos; Paulino Pérez; Daniel Gianola; Juan Burgueño; José Luis Araus; Dan Makumbi; Ravi P. Singh; Susanne Dreisigacker; Jianbing Yan; et al.Vivi AriefMarianne BanzigerHans-Joachim Braun Prediction of Genetic Values of Quantitative Traits in Plant Breeding Using Pedigree and Molecular Markers. Genetics 2010, 186, 713-724, 10.1534/genetics.110.118521.
- Elliot L. Heffner; Mark E. Sorrells; Jean-Luc Jannink; Genomic Selection for Crop Improvement. Crop Science 2009, 49, 1-12, 10.2135/cropsci2008.08.0512.
- Nicolas Heslot; Jean-Luc Jannink; Mark E. Sorrells; Perspectives for Genomic Selection Applications and Research in Plants. Crop Science 2015, 55, 1-12, 10.2135/cropsci2014.03.0249.
- Marcio P. Arruda; Patrick J. Brown; Alexander E. Lipka; Allison M. Krill; Carrie Thurber; Frederic L. Kolb; Genomic Selection for Predicting Fusarium Head Blight Resistance in a Wheat Breeding Program. The Plant Genome 2015, 8, 1-12, 10.3835/plantgenome2015.01.0003.
- J. Crossa; Paul E Oviedo Perez; John Hickey; Juan Burgueno; Leonardo Ornella; J J Ceronrojas; X. Zhang; S. Dreisigacker; R. Babu; Yun Li; et al.D G BonnettKatherine D Mathews Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity 2013, 112, 48-60, 10.1038/hdy.2013.16.
- Jesse Poland; Jeffrey Endelman; Julie Dawson; Jessica Rutkoski; Shuangye Wu; Yann Manes; Susanne Dreisigacker; José Crossa; Héctor Sánchez‐Villeda; Mark Sorrells; et al.Jean‐Luc Jannink Genomic Selection in Wheat Breeding using Genotyping‐by‐Sequencing. The Plant Genome 2012, 5, 103-113, 10.3835/plantgenome2012.06.0006.
- Jessica Rutkoski; Jared Benson; Yi Jia; Gina Brown-Guedira; Jean-Luc Jannink; Mark Sorrells; Evaluation of Genomic Prediction Methods for Fusarium Head Blight Resistance in Wheat. The Plant Genome 2012, 5, 51-61, 10.3835/plantgenome2012.02.0001.
- Elliot L. Heffner; Jean‐Luc Jannink; Mark E. Sorrells; Genomic Selection Accuracy using Multifamily Prediction Models in a Wheat Breeding Program. The Plant Genome 2011, 4, 65-75, 10.3835/plantgenome2010.12.0029.
- Deepak K. Ray; Nathaniel D. Mueller; Paul C. West; Jonathan A. Foley; Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLOS ONE 2013, 8, e66428, 10.1371/journal.pone.0066428.
- Rutkoski, J.E., Crain, J., Poland, J., Sorrells, M.E.. Genomic Selection for Crop Improvement: New Molecular Breeding Strategies for Crop Improvement; Varshney, R.K., Roorkiwal, M., Sorrells, M.E., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 99-130.
- Rex Bernardo; Jianming Yu; Prospects for Genomewide Selection for Quantitative Traits in Maize. Crop Science 2007, 47, 1082-1090, 10.2135/cropsci2006.11.0690.
- Sorrells, M.E.. Advances in Wheat Genetics: From Genome to Field; Ogihara, Y., Takumi, S., Handa, H., Eds.; Springer Japan KK: Yokohama, Japan, 2015; pp. 401-409.
- Kai Peter Voss-Fels; Mark Cooper; Ben John Hayes; Accelerating crop genetic gains with genomic selection. Theoretical and Applied Genetics 2018, 132, 669-686, 10.1007/s00122-018-3270-8.
- I. Van Den Berg; T.H.E. Meuwissen; I.M. MacLeod; M.E. Goddard; Predicting the effect of reference population on the accuracy of within, across, and multibreed genomic prediction. Journal of Dairy Science 2019, 102, 3155-3174, 10.3168/jds.2018-15231.
- Filippo M. Bassi; Alison R. Bentley; Gilles Charmet; Rodomiro Ortiz; Jose Crossa; Breeding schemes for the implementation of genomic selection in wheat ( Triticum spp . ). Plant Science 2016, 242, 23-36, 10.1016/j.plantsci.2015.08.021.
- Charlotte D. Robertsen; Rasmus L. Hjortshøj; Luc L. Janss; Genomic Selection in Cereal Breeding. Agronomy 2019, 9, 95, 10.3390/agronomy9020095.
- Julio Isidro; Jean-Luc Jannink; Deniz Akdemir; Jesse Poland; Nicolas Heslot; Mark E. Sorrells; Training set optimization under population structure in genomic selection. Theoretical and Applied Genetics 2014, 128, 145-158, 10.1007/s00122-014-2418-4.
- Adam Norman; Julian Taylor; James Edwards; Haydn Kuchel; Optimising Genomic Selection in Wheat: Effect of Marker Density, Population Size and Population Structure on Prediction Accuracy. G3 Genes|Genomes|Genetics 2018, 8, 2889-2899, 10.1534/g3.118.200311.
- Gustavo De Los Campos; John M. Hickey; Ricardo Pong-Wong; Hans D. Daetwyler; Mario P. L. Calus; Whole-Genome Regression and Prediction Methods Applied to Plant and Animal Breeding. Genetics 2013, 193, 327-345, 10.1534/genetics.112.143313.
- Elliot L. Heffner; Jean-Luc Jannink; Hiroyoshi Iwata; Edward Souza; Mark E. Sorrells; Genomic Selection Accuracy for Grain Quality Traits in Biparental Wheat Populations. Crop Science 2011, 51, 2597-2606, 10.2135/cropsci2011.05.0253.
- Guozheng Liu; Yusheng Zhao; Manje Gowda; C. Friedrich H. Longin; Jochen C. Reif; Michael F. Mette; Predicting Hybrid Performances for Quality Traits through Genomic-Assisted Approaches in Central European Wheat. PLOS ONE 2016, 11, e0158635, 10.1371/journal.pone.0158635.
- Sarah D. Battenfield; Carlos Guzmán; R. Chris Gaynor; Ravi P. Singh; Roberto J. Peña; Susanne Dreisigacker; Allan K. Fritz; Jesse A. Poland; Genomic Selection for Processing and End‐Use Quality Traits in the CIMMYT Spring Bread Wheat Breeding Program. The Plant Genome 2016, 9, 1-12, 10.3835/plantgenome2016.01.0005.
- Sebastian Michel; Christian Kummer; Martin Gallee; Jakob Hellinger; Christian Ametz; Batuhan Akgöl; Doru Epure; Franziska Löschenberger; Hermann Buerstmayr; Improving the baking quality of bread wheat by genomic selection in early generations. Theoretical and Applied Genetics 2017, 131, 477-493, 10.1007/s00122-017-2998-x.
- Mao Huang; Antonio Cabrera; Amber Hoffstetter; Carl Griffey; David Van Sanford; José Costa; Anne McKendry; Shiaoman Chao; Clay Sneller; Genomic selection for wheat traits and trait stability. Theoretical and Applied Genetics 2016, 129, 1697-1710, 10.1007/s00122-016-2733-z.
- José Crossa; Diego Jarquín; Jorge Franco; Paulino Pérez-Rodríguez; Juan Burgueño; Carolina Saint-Pierre; Prashant Vikram; Carolina Sansaloni; Cesar Petroli; Deniz Akdemir; et al.Clay SnellerMatthew ReynoldsMaria TattarisThomas PayneCarlos GuzmanRoberto J. PeñaPeter WenzlSukhwinder Singh Genomic Prediction of Gene Bank Wheat Landraces. G3 Genes|Genomes|Genetics 2016, 6, 1819-1834, 10.1534/g3.116.029637.
- Ji Yao; Dehui Zhao; Xinmin Chen; Yong Zhang; Jiankang Wang; Use of genomic selection and breeding simulation in cross prediction for improvement of yield and quality in wheat (Triticum aestivum L.). The Crop Journal 2018, 6, 353-365, 10.1016/j.cj.2018.05.003.
- B. J. Hayes; J. Panozzo; C. K. Walker; A. L. Choy; S. Kant; D. Wong; J. Tibbits; H. D. Daetwyler; S. Rochfort; M. J. Hayden; et al.G. C. Spangenberg Accelerating wheat breeding for end-use quality with multi-trait genomic predictions incorporating near infrared and nuclear magnetic resonance-derived phenotypes. Theoretical and Applied Genetics 2017, 130, 2505-2519, 10.1007/s00122-017-2972-7.
- Jemanesh K. Haile; Amidou N’Diaye; Fran Clarke; John Clarke; Ron Knox; Jessica Rutkoski; Filippo M. Bassi; Curtis J. Pozniak; Genomic selection for grain yield and quality traits in durum wheat. Molecular Breeding 2018, 38, 75, 10.1007/s11032-018-0818-x.
- Xiaowei Hu; Brett F. Carver; Carol Powers; Liuling Yan; Lan Zhu; Charles Chen; Effectiveness of Genomic Selection by Response to Selection for Winter Wheat Variety Improvement. The Plant Genome 2019, 12, 180090-15, 10.3835/plantgenome2018.11.0090.
- Hsin-Yuan Tsai; Luc L. Janss; Jeppe R. Andersen; Jihad Orabi; Jens D. Jensen; Ahmed Jahoor; Just Jensen; Genomic prediction and GWAS of yield, quality and disease-related traits in spring barley and winter wheat. Scientific Reports 2020, 10, 1-15, 10.1038/s41598-020-60203-2.
- Sebastian Michel; Christian Ametz; Huseyin Gungor; Doru Epure; Heinrich Grausgruber; Franziska Löschenberger; Hermann Buerstmayr; Genomic selection across multiple breeding cycles in applied bread wheat breeding. Theoretical and Applied Genetics 2016, 129, 1179-1189, 10.1007/s00122-016-2694-2.
- Sebastian Michel; Martin Gallee; Franziska Löschenberger; Hermann Buerstmayr; Christian Kummer; Improving the baking quality of bread wheat using rapid tests and genomics: The prediction of dough rheological parameters by gluten peak indices and genomic selection models. Journal of Cereal Science 2017, 77, 24-34, 10.1016/j.jcs.2017.07.012.
- Bettina Lado; Daniel Vázquez; Martin Quincke; Paula Silva; Ignacio Aguilar; Lucia Gutiérrez; Resource allocation optimization with multi-trait genomic prediction for bread wheat (Triticum aestivum L.) baking quality. Theoretical and Applied Genetics 2018, 131, 2719-2731, 10.1007/s00122-018-3186-3.
- Sebastian Michel; Franziska Löschenberger; Christian Ametz; Bernadette Pachler; Ellen Sparry; Hermann Bürstmayr; Combining grain yield, protein content and protein quality by multi-trait genomic selection in bread wheat. Theoretical and Applied Genetics 2019, 132, 2767-2780, 10.1007/s00122-019-03386-1.
- Kristensen, P.S.; Jahoor, A.; Andersen, J.R.; Orabi, J.; Janss, L.; Jensen, J.; Multi-Trait and Trait-Assisted Genomic Prediction of Winter Wheat Quality Traits Using Advanced Lines from Four Breeding Cycles. Crop Breeding, Genetics and Genomics 2019, 1, e1900010, 10.20900/cbgg20190010.
- Maria Itria Ibba; Jose Crossa; Osval A. Montesinos‐López; Abelardo Montesinos‐López; Philomin Juliana; Carlos Guzman; Emily Delorean; Susanne Dreisigacker; Jesse Poland; Genome‐based prediction of multiple wheat quality traits in multiple years. The Plant Genome 2020, 13, e20034, 10.1002/tpg2.20034.
- Maria Itria Ibba; Jose Crossa; Osval A. Montesinos‐López; Abelardo Montesinos‐López; Philomin Juliana; Carlos Guzman; Emily Delorean; Susanne Dreisigacker; Jesse Poland; Genome‐based prediction of multiple wheat quality traits in multiple years. The Plant Genome 2020, 13, e20034, 10.1002/tpg2.20034.
 Encyclopedia
Encyclopedia


