A major challenge to the papaya industry is inconsistency in fruit quality and, in particular, flavour, which is a complex trait that comprises taste perception in the mouth (sweetness, acidity, or bitterness) and aroma produced by several volatile compounds. Current commercial varieties vary greatly in their taste, likely due to historical prioritised selection for fruit appearance as well as large environmental effects. Therefore, it is important to better understand the genetic and biochemical mechanisms and biosynthesis pathways underpinning preferable flavour in order to select and breed for better tasting new commercial papaya varieties. As an initial step, objectively measurable standards of the compound profiles that provide papaya’s taste and aroma, together with ‘mouth feel’, are required. This review presents an overview of the approaches to characterise the flavour profiles of papaya through sugar component determination, volatile compound detection, sensory panel testing, as well as genomics-based studies to identify the papaya flavour.
- papaya breeding
- flavour profiling
- biosynthesis pathways
- gene identification
1. Introduction
Papaya ( Carica papaya L.) is one of the top five most commonly grown tropical fruit crops throughout tropical and subtropical regions worldwide, including in Australia, Hawaii, and Southeast Asia [1]. Papaya fruit is juicy with a sweet flavour and the ripe fruit is rich in vitamins A and C, folate, as well as calcium [2]. The fruit is valued for its nutritional status and is usually eaten raw, whereas unripe green fruit can be eaten both raw and cooked, for example, in green papaya salad. In addition, unripe papaya is a source of papain, an endolytic plant cysteine protease that plays a crucial role in many vital biological processes in all living organisms and that has been used in meat tenderising for thousands of years [3][4].
Due to its high productivity, nutritional value, and functionality, papaya has become an important commercial fruit crop worldwide. The global production of papaya for the past twenty years has steadily increased, mainly because of increased production in India and demand by the United States, reaching a peak in 2016 of 13.09 million tonnes. In 2018, 60.9% of the world’s total papaya production was in three countries: India (138 thousand ha and 5.99 million tonnes), Brazil (27.2 thousand ha and 1.06 million tonnes), and Mexico (18 thousand ha and 1.04 million tonnes) [5].
Although great gains have been made in increasing fruit yield, there has generally not been a simultaneous improvement in flavour quality. This has likely led to reduced market uptake, hence recent breeding has focused on improving fruit and flavour quality traits with the intention to expand the market [6]. Fruit quality comprises several important factors including flavour, nutrition, appearance, texture, and postharvest processing. Flavour is a complex trait that includes taste perception in the mouth (sweetness, acidity, and/or bitterness) and aroma, which is produced by several volatile compounds [7]. Therefore, the combination of both mouth perception as well as amounts and ratios of volatile compounds present in the flesh plays a major role in determining the perception and acceptability of papaya flavour by consumers. Together, these considerations are important in strategic breeding, branding, and marketing of premium papaya cultivars to align with consumer acceptance and demand. To achieve this, objective standards of good taste and aroma must be set. Additionally, molecular markers for detecting desirable fruit traits at an early stage of papaya growth are needed for targeted genomics-assisted breeding strategies. The following is a review of the key factors and current knowledge in papaya fruit flavour quality and the considerations for the strategic breeding of the flavour of preference.
2. Papaya Flavour
Many factors, including the biochemical and environmental contributions to flavour profiles, how these are perceived, and the suitable eating stage, must be understood to successfully breed papaya and improve its flavour. Fruit flavour comprises sugars, acids, and volatile components [8]. The combination of a consumer’s mouth perception together with an understanding of the preferred acidity, sweetness, and known amounts and ratios of specific volatile compounds can be used to develop a tool to differentiate and select for flavour types or profiles. In this review section, sugar accumulation and volatile compounds, as well as the genes involved in fruit flavour metabolism pathways in papaya, will be discussed.
Papaya sweetness is contributed to by three main soluble sugars: glucose, fructose, and sucrose [9][10][11][12]. Accordingly, for papaya sweetness evaluation, it is essential to quantify total sugar content and the ratio of each type of sugar in that total [12][13]. During papaya fruit development, sugar accumulation initiates after seed maturation with the increasing activity of sucrose synthase (SS), and glucose is the major soluble sugar [9][14]. The metabolic pathway of sugar metabolism and associated enzymes in papaya is shown in Figure 1 .
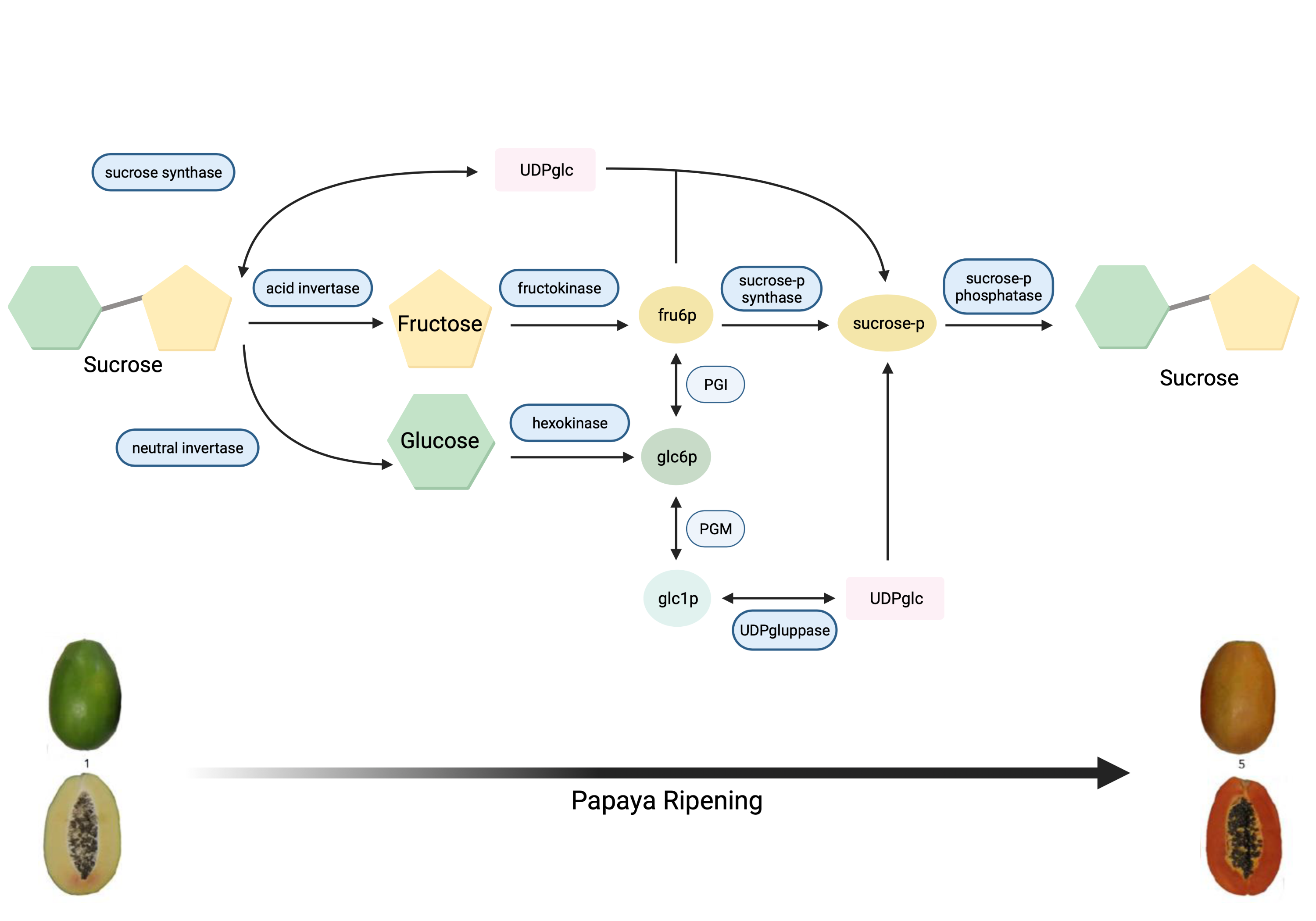 Figure 1. Metabolic pathway of sugar metabolism in papaya fruit, indicating enzyme reactions.
Figure 1. Metabolic pathway of sugar metabolism in papaya fruit, indicating enzyme reactions.
Invertases were also involved in the accumulation of glucose and fructose in ripe papaya fruit variety ‘Golden’ [15]. Sucrose phosphate synthase (SPS) plays an important role in metabolising neutral sugars, especially mannose from cell walls for the continuous synthesis of sucrose and galactose, the main source of carbon during the synthesis of sucrose [13]. Galactose is metabolised rapidly under high SPS activities, liberating simple sugars, and contributes to sweetness [16]. In addition, several quantitative trait loci (QTLs) for flesh sweetness have been detected in papaya varieties ‘RB2’ and ‘Sunrise Solo’, which are associated with the SS gene family [17][18]. Nantawan et al. [17] determined that sucrose was the main sugar in these two papaya varieties at the ripening stage, contributing 40–60% of the total sugar.
Almost 400 volatiles were originally identified in papaya [19] using a range of separation techniques such as headspace, gas chromatography, odour olfactometry, and mass spectrometry [8][20][21][22]. Based on these previous studies, the characteristic aroma of papaya fruit is proposed to be produced by combinations of alcohols, esters, aldehydes, and sulphur compounds [23].
3. The Genomics of Fruit Flavour
The genes that contribute to fruit flavour are involved in the production of sugars, acids, and volatile components, which are also involved in other primary and secondary metabolic pathways [6]. Therefore, genomics-based studies to identify the genes controlling flavour and the subsequent development of selective molecular markers have focused on understanding those within the sugar and volatile syntheses pathways [6][17][24].
To select papaya genotypes with high sugar content phenotypes, several of the functional genes related to sugar synthesis and accumulation pathways have been identified [17][18]. As mentioned in the previous section, invertase, SPS, and SS are the key enzymes involved in papaya sugar production throughout the ripening stage [25][26][9][27][10]. Genes involved in the production of these key enzymes have been investigated and identified in a variety of fruits including tomato [28], pineapple [29], apple [30], and papaya [18]. In some fruits, such as tomato [31], citrus [32], and sugarcane [33], SPS activity is directly linked to sucrose accumulation during the fruit maturation stage, but the detailed mechanisms are still unknown. Meanwhile, in other fruits, such as grape berries [34] and pineapple [29], the mechanism for sucrose production is more complicated and regulated by multiple genes.
Several genes have been identified for the improvement of fruit sweetness in papaya, including nine predicted to control sugar synthesis and sugar transportation [35]. Among these, two were papaya cell wall invertase genes ( CpCWINV1 and CpCWINV2 ), which have the Glycohydrolase Family 32 (GH32) domain and are responsible for exporting sucrose from the phloem to the cell as well as hydrolysing sucrose into fructose and glucose. A further four were SS genes ( CpSUS1 to CpSUS4 ) and all had sucrose synthase and glycosyl transferase (GT4) domains, which catalyse the reversible conversion of sucrose and UDP to UDP-glucose and fructose. Another three were SPS genes ( CpSPS1 , CpSPS2 , CpSPS3 ) and all had GT4 and SPS domains. These genes were selected as sweetness candidate genes and identified in two papaya genotypes ‘RB2′ and ‘Sunrise Solo’ by Nantawan et al. [18]. Higher expression levels of cpSPS1 , cpSPS2 , cpSPS3 , cpSPS4 , cpCWINV1 , and cpAVIN2 were observed in ‘Sunrise Solo’, the genotype with a high sugar content ( Figure 2 ). This indicated major putative roles of these genes in sugar synthesis in papaya [18]. A sugar transporter gene ( AT3G05165 ) was also discovered in ripe papaya (cv. ‘Golden’) by Fabi et al. in 2012 [36]. Additionally, nine putative enzymes associated with sugars/sugar alcohols with unigenes were identified in ripe papaya (cv. ‘Eksotika’) by using mRNA paired-end sequencing [37]. These were α-galactosidase, α-glucosidase, myo-inositol monophosphatase, β-fructofuranosidase, xylose isomerase, fructose biphosphatase, fructose biphosphate aldose, ribose 5-phosphate isomerase, arabinose kinase, and β-glucosidase.
Volatiles can influence the perception of sweetness and vice versa [39]. Fruit-related volatiles are mainly derived from three different secondary metabolic pathways; (1) the isoprenoid biosynthesis pathway that produces mono- and sesquiterpenes, (2) the shikimic acid aromatic amino acid biosynthesis pathway that produces phenylpropanoids and benzenoid fragrances, and (3) the acyl lipid catabolism pathway that forms short branched-chain aldehydes, alcohols, esters, and ketones. The genes within these pathways represent targets for studies focused on uncovering differential sequence identification, differential expression, and the development of biomarkers for preferred volatile selections [24]. The key flavour volatile genes are generally divided into two classes: (1) those encoding enzymes responsible for the synthesis of the end product and (2) those encoding factors that regulate the synthetic pathways [24]. In papaya, the main contributors to aroma are methyl and ethyl ester derivatives of lipid catabolism [40]. Accordingly, 14 papaya sequences were predicted to be associated with enzymes involved in fruit volatile biosynthesis. Among these, five were highly similar to enzymes in the acyl lipid catabolism pathway, and two were enzymes that encoded the final steps of the fatty acid degradation to acetyl-coenzyme A pathway: Abnormal inflorescence meristem 1/fatty acid multifunctional protein (AIM1) and 3-ketoacyl-CoA thiolase via the β-oxidation pathway [40]. An additional two genes ( CpPALDS2 and CpPALDR1 ) were involved in volatile production from phenylalanine as identified by [35].
4. Gene Identification and Functional Validation
The identification and characterisation of gene sequences related to papaya sweetness and volatile compounds are important for future breeding, branding, and marketing of premium papaya cultivars to align with consumer acceptance and demand. The first step towards this is the identification and functional validation of these genes/sequences within current papaya commercial varieties and advanced breeding lines.
Therefore, RNA-Seq and qPCR analyses together with biochemical analyses may be a suitable combined approach for uncovering the genomic components of papaya flavour. An immediate limitation to RNA-Seq analysis in papaya, however, is the lack of a dense coverage and fully annotated papaya reference genome. This is surprising since the papaya genome size is relatively small (372 Mbp) when compared with other plants such as banana (875 Mbp), plum (883 Mbp), and avocado (883 Mbp) [41]. The first draft genome of papaya was released by Ming et al. (2008), who sequenced the transgenic variety ‘SunUp’, the first commercially available and virus-resistant transgenic papaya, by using Sanger sequencing technology followed by BAC-end sequences as well as physical and genetic maps [42]. To date, this stands as the most comprehensive published papaya reference genome ( http://www.plantgdb.org/CpGDB/ , accessed on 29 May 2021), yielding 1.6 million high quality reads from a total of 2.8 million whole genome shotgun (WGS) sequencing reads assembled into 271Mb contigs and representing around 75% of the papaya genome. A total of 24,746 genes were accurately annotated from this following the TIGR Eukaryotic Annotation Pipeline [43]. This was a 11–20% lower gene count than detected in Arabidopsis [44] and 34% less than in rice [45]. A denser and better annotated genome assembly is much needed for future advanced papaya genomics studies.
CRISPR/Cas9 vector systems have been widely used in the efficient editing of plant genomes including for gene knockout, gene knock in, and the suppression of virus infection [46]. In rice, Shimatani et al. [47] induced three multiple-herbicide-resistance point mutations (C287T, G590, and W483), which conferred resistance to the herbicide imazamox (IMZ). By mutating these genes in rice, they were able to induce 3.41% IMZ tolerance. In papaya, CRISPR/Cas9-mediated mutations of S-genes have been developed to enable resistance to papaya ringspot virus (PRSV) [48][49]. However, the CRISPR/Cas9 approach requires a robust transformation system and works best when a clear and single target is identified.
Conversely, flavour is a very complex multigenic trait, comprising numerous reactions mediated by multiple enzymes and genes. Flavour is also likely influenced by several physical and environmental factors, which can lead to qualitative and quantitative changes in biochemical component production [50].
References
- Carr, M.K.V. The water relations and irrigation requirements of papaya (Carica papaya L.): A review. Exp. Agric. 2014, 50, 270–283.
- Nutrition. Papaya Aust. Available online: https://australianpapaya.com.au/about/nutrition/ (accessed on 29 May 2021).
- Tsuge, H.; Nishimura, T.; Tada, Y.; Asao, T.; Turk, D.; Turk, V.; Katunuma, N. Inhibition Mechanism of Cathepsin L-Specific Inhibitors Based on the Crystal Structure of Papain–CLIK148 Complex. Biochem. Biophys. Res. Commun. 1999, 266, 411–416.
- Olmoss, A. Papain, a Plant Enzyme of Biological Importance: A Review. Am. J. Biochem. Biotechnol. 2012, 8, 99–104.
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 29 May 2021).
- Klee, H.J. Improving the Flavor of Fresh Fruits: Genomics, Biochemistry, and Biotechnology. New Phytol. 2010, 187, 44–56.
- El Hadi, M.A.M.; Zhang, F.-J.; Wu, F.-F.; Zhou, C.-H.; Tao, J. Advances in Fruit Aroma Volatile Research. Molecules 2013, 18, 8200–8229.
- Wijaya, C.H.; Chen, F. Flavour of Papaya (Carica papaya L.) fruit. BIOTROPIA Southeast Asian J. Trop. Biol. 2013, 20, 1.
- Zhou, L.; Paull, R.E. Sucrose Metabolism During Papaya (Carica papaya) Fruit Growth and Ripening. J. Am. Soc. Hortic. Sci. 2001, 126, 351–357.
- Paull, R.E.; Gross, K.; Qiu, Y. Changes in Papaya Cell Walls during Fruit Ripening. Postharvest Biol. Technol. 1999, 16, 79–89.
- Gomez, L.; Bancel, D.; Rubio, E.; Vercambre, G. The Microplate Reader: An Efficient Tool for the Separate Enzymatic Analysis of Sugars in Plant Tissues—Validation of a Micro-Method. J. Sci. Food Agric. 2007, 87, 1893–1905.
- Othman, R.; Nuraziyan, A. Fruit-Specific Expression of Papaya Subtilase Gene. J. Plant Physiol. 2010, 167, 131–137.
- Gomez, M.; Lajolo, F.; Cordenunsi, B. Evolution of Soluble Sugars During Ripening of Papaya Fruit and Its Relation to Sweet Taste. J. Food Sci. 2002, 67, 442–447.
- Fabi, J.P.; Peroni, F.H.G. Papaya, Mango and Guava Fruit Metabolism during Ripening: Postharvest Changes Affecting Tropical Fruit Nutritional Content and Quality. Fresh Prod. 2010, 4, 56–64.
- Nogueira, S.B.; Labate, C.A.; Gozzo, F.C.; Pilau, E.J.; Lajolo, F.M.; Oliveira do Nascimento, J.R. Proteomic Analysis of Papaya Fruit Ripening Using 2DE-DIGE. J. Proteomics 2012, 75, 1428–1439.
- Yao, B.N.; Tano, K.; Konan, H.K.; Bédié, G.K.; Oulé, M.K.; Koffi-Nevry, R.; Arul, J. The Role of Hydrolases in the Loss of Firmness and of the Changes in Sugar Content during the Post-Harvest Maturation of Carica papaya L. Var Solo 8. J. Food Sci. Technol. 2014, 51, 3309–3316.
- Nantawan, U.; Kanchana-udomkan, C.; Bar, I.; Ford, R. Linkage Mapping and Quantitative Trait Loci Analysis of Sweetness and Other Fruit Quality Traits in Papaya. BMC Plant. Biol. 2019, 19, 449.
- Nantawan, U.; Kanchana-udomkan, C.; Drew, R.; Ford, R. Identification of Genes Related to Sugar Content in Carica papaya L.: Differential Expression and Candidate Marker Development. In Proceedings of the Acta Horticulturae; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2018; pp. 129–136.
- Pino, J.A.; Almora, K.; Marbot, R. Volatile Components of Papaya (Carica papaya L., Maradol Variety) Fruit. Flavour Fragr. J. 2003, 18, 492–496.
- Flath, R.A.; Forrey, R.R. Volatile Components of Papaya (Carica papaya L., Solo Variety). J. Agric. Food Chem. 1977, 25, 103–109.
- Lieb, V.M.; Esquivel, P.; Cubero Castillo, E.; Carle, R.; Steingass, C.B. GC–MS Profiling, Descriptive Sensory Analysis, and Consumer Acceptance of Costa Rican Papaya (Carica papaya L.) Fruit Purees. Food Chem. 2018, 248, 238–246.
- Ulrich, D.; Wijaya, C. Volatile Patterns of Different Papaya (Carica papaya L.) Varieties. J. Appl. Bot. Food Qual. 2010, 83, 128–132.
- Berger, R.G. Flavours and Fragrances: Chemistry, Bioprocessing and Sustainability; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007; ISBN 978-3-540-49339-6.
- Chambers, A.H.; Pillet, J.; Plotto, A.; Bai, J.; Whitaker, V.M.; Folta, K.M. Identification of a Strawberry Flavor Gene Candidate using an Integrated Genetic-Genomic-Analytical Chemistry Approach. BMC Genomics 2014, 15, 1–15.
- Yamaki, S. Metabolism and Accumulation of Sugars Translocated to Fruit and Their Regulation. J. Jpn. Soc. Hortic. Sci. 2010, 79, 1–15.
- Hubbard, N.L.; Pharr, D.M.; Huber, S.C. Sucrose Phosphate Synthase and Other Sucrose Metabolizing Enzymes in Fruits of Various Species. Physiol. Plant. 1991, 82, 191–196.
- Patrick, J.W.; Botha, F.C.; Birch, R.G. Metabolic Engineering of Sugars and Simple Sugar Derivatives in Plants. Plant. Biotechnol. J. 2013, 11, 142–156.
- Sagor, G.H.M.; Berberich, T.; Tanaka, S.; Nishiyama, M.; Kanayama, Y.; Kojima, S.; Muramoto, K.; Kusano, T. A Novel Strategy to Produce Sweeter Tomato Fruits with High Sugar Contents by Fruit-Specific Expression of a Single BZIP Transcription Factor Gene. Plant. Biotechnol. J. 2016, 14, 1116–1126.
- Zhang, X.; Du, L.; Xie, J.; Dou, M.; Sun, G. Cloning and Expression of Pineapple Sucrose- Phosphate Synthase Gene during Fruit Development. Afr. J. Biotechnol. 2010, 9, 8296–8303.
- Li, M.; Feng, F.; Cheng, L. Expression Patterns of Genes Involved in Sugar Metabolism and Accumulation during Apple Fruit Development. PLoS ONE 2012, 7, e33055.
- Dali, N.; Michaud, D.; Yelle, S. Evidence for the Involvement of Sucrose Phosphate Synthase in the Pathway of Sugar Accumulation in Sucrose-Accumulating Tomato Fruits 1. Plant Physiol. 1992, 99, 434–438.
- Komatsu, A.; Takanokura, Y.; Moriguchi, T.; Omura, M.; Akihama, T. Differential Expression of Three Sucrose-Phosphate Synthase Isoforms during Sucrose Accumulation in Citrus Fruits (Citrus Unshiu Marc.). Plant Sci. 1999, 140, 169–178.
- Lingle, S.E. Sugar Metabolism during Growth and Development in Sugarcane Internodes. Crop Sci. 1999, 39, 480.
- Davies, C.; Robinson, S.P. Sugar Accumulation in Grape Berries (Cloning of Two Putative Vacuolar Invertase CDNAs and Their Expression in Grapevine Tissues). Plant Physiol. 1996, 111, 275–283.
- Paull, R.E.; Irikura, B.; Wu, P.; Turano, H.; Chen, N.J.; Blas, A.; Fellman, J.K.; Gschwend, A.R.; Wai, C.M.; Yu, Q.; et al. Fruit Development, Ripening and Quality Related Genes in the Papaya Genome. Trop. Plant Biol. 2008, 1, 246–277.
- Fabi, J.P.; Seymour, G.B.; Graham, N.S.; Broadley, M.R.; May, S.T.; Lajolo, F.M.; Cordenunsi, B.R.; Oliveira do Nascimento, J.R. Analysis of Ripening-Related Gene Expression in Papaya Using an Arabidopsis-Based Microarray. BMC Plant Biol. 2012, 12, 242.
- Sanimah, S.; Maheswary, V.; Sarip, J.; Qistina, O.N.; Vasanthi, S. Identification of Phytochemicals and the Associated Genes in Eksotika Papaya at Ripening Index 5 Using Functional Genomics. J. Trop. Agric. Food Sci. 2013, 41, 283–308.
- KEGG for Integration and Interpretation of Large-Scale Molecular Data Sets|Nucleic Acids Research|Oxford Academic. Available online: academic.oup.com/nar/article/40/D1/D109/2903713?login=true (accessed on 10 April 2021).
- Baldwin, E.A.; Goodner, K.; Plotto, A. Interaction of Volatiles, Sugars, and Acids on Perception of Tomato Aroma and Flavor Descriptors. J. Food Sci. 2008, 73, S294–S307.
- Devitt, L.C.; Sawbridge, T.; Holton, T.A.; Mitchelson, K.; Dietzgen, R.G. Discovery of Genes Associated with Fruit Ripening in Carica papaya Using Expressed Sequence Tags. Plant Sci. 2006, 170, 356–363.
- Arumuganathan, K.; Earle, E.D. Nuclear DNA Content of Some Important Plant Species. Plant Mol. Biol. Report. 1991, 9, 208–218.
- Ming, R.; Hou, S.; Feng, Y.; Yu, Q.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.T.; et al. The Draft Genome of the Transgenic Tropical Fruit Tree Papaya (Carica papaya Linnaeus). Nature 2008, 452, 991–996.
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource: Improvements and New Features. Nucleic Acids Res. 2007, 35, D883–D887.
- Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; et al. Genome Sequencing and Analysis of the Model Grass Brachypodium Distachyon. Nature 2010, 463, 763–768.
- Sasaki, T. International Rice Genome Sequencing Project The Map-Based Sequence of the Rice Genome. Nature 2005, 436, 793–800.
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y.-G. CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications. Mol. Plant 2016, 9, 961–974.
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted Base Editing in Rice and Tomato Using a CRISPR-Cas9 Cytidine Deaminase Fusion. Nat. Biotechnol. 2017, 35, 441–443.
- Gumtow, R.; Wu, D.; Uchida, J.; Tian, M. A Phytophthora Palmivora Extracellular Cystatin-Like Protease Inhibitor Targets Papain to Contribute to Virulence on Papaya. Mol. Plant-Microbe Interact. 2017, 31, 363–373.
- Hamim, I.; Borth, W.B.; Marquez, J.; Green, J.C.; Melzer, M.J.; Hu, J.S. Transgene-Mediated Resistance to Papaya Ringspot Virus: Challenges and Solutions. Phytoparasitica 2018, 46, 1–18.
- Tikunov, Y.M.; Roohanitaziani, R.; Meijer-Dekens, F.; Molthoff, J.; Paulo, J.; Finkers, R.; Capel, I.; Moreno, F.C.; Maliepaard, C.; Vries, M.N.; et al. The Genetic and Functional Analysis of Flavor in Commercial Tomato: The FLORAL4 Gene Underlies a QTL for Floral Aroma Volatiles in Tomato Fruit. Plant J. 2020, 103, 1189–1204.
 Encyclopedia
Encyclopedia


