Rapid breakthroughs in nucleic acid nanotechnology have always driven the creation of nano-assemblies with programmable design, potent functionality, good biocompatibility, and remarkable biosafety during the last few decades. Researchers are constantly looking for more powerful techniques that provide enhanced accuracy with greater resolution. The self-assembly of rationally designed nanostructures is now possible because of bottom-up structural nucleic acid (DNA and RNA) nanotechnology, notably DNA origami. Because DNA origami nanostructures can be organized precisely with nanoscale accuracy, they serve as a solid foundation for the exact arrangement of other functional materials for use in a number of applications in structural biology, biophysics, renewable energy, photonics, electronics, medicine, etc. DNA origami facilitates the creation of next-generation drug vectors to help in the solving of the rising demand on disease detection and therapy, as well as other biomedicine-related strategies in the real world. These DNA nanostructures, generated using Watson–Crick base pairing, exhibit a wide variety of properties, including great adaptability, precise programmability, and exceptionally low cytotoxicity in vitro and in vivo.
- DNA origami
- DNA nanostructures
- self-assembly
- drug encapsulation
1. Introduction
2. Overview and Structural Features of DNA Origami Nanostructures
3. Synthesis and Assembly of DNA Origami Nanostructures
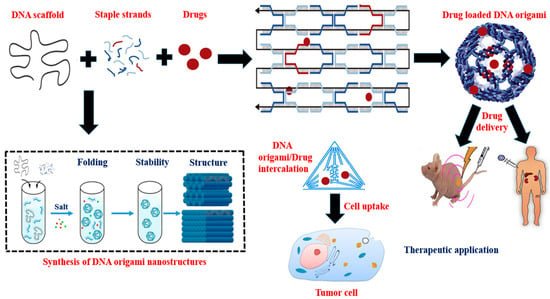
4. DNA-Origami-Based Approaches and Therapeutic Strategies for Targeted Drug Delivery
The anti-inflammatory, antioxidant, toxic, anti-cancer, and immunomodulatory properties of quercetin (flavonoids) demonstrate its potential therapeutic value. Despite its many positive effects on human health, quercetin has some drawbacks, including its hydrophobic nature, low bioavailability, poor solubility, and poor permeability. Quercetin is encapsulated in DNA origami to improve its solubility and absorption in order to overcome some of its drawbacks [40].
A typical flavonoid is luteolin, a 3',4',5,7-tetrahydroxyflavone that may be found in a wide range of plants, including fruits, vegetables, and medicinal herbs. luteolin- rich plants have been utilized in Chinese traditional medicine to treat a range of diseases, including cancer, inflammatory conditions, and hypertension. Luteolin contains a range of biological effects, such as anti-allergy, anti-inflammation and anticancer. It may function biochemically as either an antioxidant or a pro-oxidant. The biological effects of luteolin could be related functionally. For instance, its anti-inflammatory activity could be connected to its anti-cancer effect. The stimulation of apoptosis and the prevention of cell growth, metastasis, and angiogenesis are two of luteolin's anticancer properties. Other drawbacks include its hydrophobic nature, limited bioavailability, poor permeability, and poor solubility. Luteolin is encapsulated in DNA origami to improve its bioavailability and solubility in order to overcome some of its drawbacks [41].
The well-known antibiotic actinomycin-D, which is intercalated in DNA origami and has significant antibacterial and anticancer action, belongs to the actinomycin group. Its cytotoxic and antitumor effects are caused by a number of mechanisms, many of which are linked to DNA functioning, which inhibits the production of RNA and, in turn, protein synthesis. The two main mechanisms are intercalation to DNA and stabilization of topoisomerases I and II cleavable complexes with DNA, in which a polypeptide lactone ring occupies a position in the minor groove of the DNA helix and a phenoxazone ring localizes between GpC base pair sequence in DNA, or the drug penetrates to a location in the DNA structure where topoisomerase binds with DNA, respectively. In addition, it has been hypothesized that actinomycin D's sluggish dissociation from DNA complexes, photodynamic activity, free radical generation, and other biochemical impacts of activity may be significant determinants of this drug’s biological action [42].5. Challenges
[A] Cost: One of the most difficult obstacles standing in the way of the practical uses of DNA origami as a medication delivery system is cost. At a synthesis scale of roughly 10 nmol, staple strands of a 7,000 bp origami structure are commonly available for purchase for several hundred dollars. The true cost of individual DNA origami design would be significantly greater if other expenses like scaffold DNA, oligonucleotide functionalization, and origami purification were taken into consideration. Therefore, it is vitally necessary to develop cost-effective scaffold and staple DNA synthesis techniques. Given that basic DNA tile structures have previously been generated in vivo, it may be possible to resolve this problem by producing DNA strands or perhaps whole origami structures in vivo [55].
[B] In vitro and in vivo stability: Another problem that has to be solved is the stability of DNA origami in vitro and in vivo. High quantities of cationic ions (such as Mg2+ and Na+) are necessary to neutralize the negative charge of the DNA backbone and maintain DNA origami structures because of the extraordinarily dense packing of DNA duplexes in these nanostructures. The amounts of cationic ions in typical physiological solutions (such PBS and medium) are insufficient to stabilize DNA origami constructs. The stability of DNA origami in fluids that simulate physiological circumstances has been tested in several experiments. Less dense things, such as wire-frame origami creations, have been discovered to be more stable in cation-depleted fluids [13][56][57].
[C] Immune response: Exogenous DNA insertion carries a number of risks, including long-term integration into the genome, induction of a strong immune response, sequence-specific interference with mRNAs or microRNAs that results in undesired gene regulation. One possible answer to these issues is the chemical alteration of the fundamental DNA strands, such as the insertion of modified phosphoramidites or post-synthetic modification to make them physiologically inactive [58].
[D] Design: Further simplified and automated design platforms need to be developed, especially for researchers outside the DNA nanotechnology field.
[E] Scale up: The size of discrete origami structures is typically constrained within 100 nm because of the length of M13 scaffold DNA, and thus alternative strategies need to be developed for size expansion.
[F] Chemical functionality: DNA is a relatively chemically inert biomolecule, and thus facile methods for adding a wide variety of functionalities needs further development. Current methods for introducing additional reactivity, through the introduction of alternative nucleotides during synthesis or post-synthetic modifications, provide a good starting point but can be cost prohibitive.
[G] Defects: DNA origami structures contain assembly defects, which can hinder heteroelement or therapeutic incorporation. Optimizing structural designs (e.g., crossover pattern and staple length) and assembly conditions (e.g., Mg2+ concentration and a thermal annealing protocol) can help to minimize structural defects. Straightforward methods should also be developed to allow convenient examination of structural quality.
6. Future Perspectives
[A] Predictable and well-defined structure: It is generally known that object size and form can affect how cells internalize substances. Given the ease and flexibility with which DNA origami nanostructures of various sizes and forms may be designed and produced This adaptability gives us a great chance to experiment with different structures as drug carriers, and there is potential to optimize a number of different factors for cellular uptake using the same uniform material [59].
[B] Stability: An important criterion is how stable DNA nanostructures are in a physiological environment. It has been demonstrated that in nuclease-containing circumstances, DNA origami are more stable than ssDNA and regular DNA duplexes. This stability may be caused by the fact that the DNA origami's odd forms and structures have physical complexity that make them difficult for nucleases to access and use. It has been demonstrated that various DNA origami nanostructures may survive for 12 hours in cell lysates at room temperature without deteriorating [43].
[C] Drug loading and release: The flexibility of DNA origami nanostructures' drug loading and release properties makes them useful for designing the structural elements of nanocarriers. Unmethylated cytosine-phosphate-guanine (CpG) sequences have been employed as a model cargo and have been covalently attached to DNA nanocarriers in order to elicit an immunological response. A Fab fragment, AuNPs, and active enzymes have all been reported to be contained inside a DNA origami nanostructure's hollow. The cargo and enzymes were able to be more stable, catalytically active, and resistant to protease digestion because to these DNA origami nanostructures, according to the data [50][60][61].
[D] Cellular internalization: It has been demonstrated that DNA origami nanostructures with larger sizes and stronger compactness enable more effective internalization than structures with smaller compactness or isolated ssDNA [60]. DNA origami nanostructures have been altered with targeted ligands, such as folate, cell-penetrating proteins, and transferrin, to increase the efficiency of cellular absorption [62]. Additionally, improved permeability and retention (EPR) effects were seen in DNA origami nanostructures. After an intravenous injection into tumor-bearing mice, the passive accumulation of DNA origami in the three distinct forms of triangle, rectangle, and tube was examined using QD labelling. It was discovered that 24 h after injection, the triangles accumulated at the tumor site at greater quantities than the tubular nanostructures [27].
[E] Therapeutic efficacy: High loading, minimal cytotoxicity, perfect stability, and releasing capability of nanocarriers all contribute to high effectiveness in cancer therapy. Numerous research shown that DNA origami nanostructures improved anticancer functions and got around drug resistance. Doxorubicin-infused triangular and tubular DNA origami nanostructures, according to Jiang and colleagues, boosted the apoptosis of doxorubicin-resistant breast cancer [63]. It has been suggested that DNA nanocarriers can lessen the negative effects of chemotherapy. When compared to mice in the free drug group, animals treated with doxorubicin-containing DNA triangles efficiently reduced tumor growth while causing minimal weight loss, demonstrating that these DNA nanocarriers were less harmful than free-drug mice [27].
[F] Photodynamic therapy: In photodynamic therapy (PDT), cancer cells are killed by combining light with photosensitizers. There are several photosensitizers for PDT, including silicon phthalocyanine Pc 4, aminolevulinic acid, and porphyrins. Some medicines, however, have drawbacks such as slow absorption, quick clearance, and poor solubility, which therefore lead to insufficient therapeutic effectiveness. Additionally, DNA origami nanostructures have been applied in PDT as nanocarriers of photosensitizers [64].
[G] Further investigations: To completely understand the stability issue with DNA origami constructs, more research is required. Although the effective cell entry of DNA origami structures has been experimentally confirmed, the precise endocytosis process has not yet been identified through rigorous mechanistic research. Another difficult obstacle is that every research has indicated that DNA origami constructs eventually make their way to lysosomes for digestion. DNA origami vehicles may be required to escape from the lysosome in order to facilitate effective cargo release into the cytosol. Potential tactics include conjugating functional molecules onto DNA origami to promote lysosomal escape or utilizing targeted ligands to start absorption via a non-lysosomal route. Before any clinical drug-delivery applications, a deeper comprehension of the pharmacokinetics and pharmacodynamics of DNA origami constructs in vivo is also required [65].
7. Conclusion
DNA origami has good drug targeting capacity and lower drug toxicity, so nanomedicine has grown at an exponential rate. It makes use of interactions in which nanotechnological materials and biological systems interface with one other to improve delivery performance. Since its start ten years ago, DNA origami has made incredible progress toward a variety of applications. This paper covers several approaches currently utilized for the construction of DNA origami nanostructures. The use of these DNA nanostructures with well-defined parameters for accurate control in the delivery of drug and gene therapy is also explored. The researchers pointed out several drugs that can be encapsulated with DNA origami and concluded the prospectives and challenges of DNA origami. While DNA origami-based nanotechnology has great potential for precise nanomedicine, but it is still in its early stages. Prior to clinical translation, certain critical obstacles must be addressed. (I) Unclear operating mechanism. Although DNA nanostructures have been explored for drug delivery, more research into the mechanics of transfection is desperately required since the real mechanism of uptake and how crucial parameters such as size and shape impact uptake are still unknown. (II) Further testing of safety profile. DNA, being a naturally biocompatible and biodegradable polymer, performs quite well in some types of cells and in mice. In these preliminary trials, there was no antibody reaction against DNA nanostructures. Nevertheless, considering the complexities of the human body, the impact of particle physicochemical characteristics on renal systems, and the doubtful but potentially harmful genome recombination, more research on DNA origami nanostructures in different types of organs is required before they can be used in clinical settings. The practical implementation of DNA origami nanostructures in vivo would be driven by additional mechanistic research of the fate of DNA nanostructures in vivo and the development of effective ways to reduce interruption from the physiological environment. A straightforward method for fabricating large-scale DNA origami nanostructures should substantially facilitate their development and translation for commercial, clinical and larger-scale research implications. Moreover, advancement in synthesizing DNA origami components and associated assemblies above the micrometer scale is rarely documented; this barrier should be eliminated in order to achieve large-scale applications in electronics, catalysis, and other fields. In this context, significant efforts should be undertaken to overcome such issues in order to assist the growth of the DNA origami techniques and DNA origami-based nanofabrication, which have the potential to radically alter multidisciplinary disciplines and manufacturing processes. Researchers believe that with further advancement in drug delivery system and resolution of scalability difficulties, DNA origami-based nanotechnology can bring a new notion into carrier systems and provide beneficial clinical results. Recent discoveries in the design and implementation of DNA origami nanostructures demonstrate that there is substantial potential for improvement, which should lead to significant applications for these nanostructures in material sciences and healthcare. This topic will undoubtedly attract multidisciplinary research efforts among chemists, biologists, doctors, and bioengineers, and interesting new discoveries will emerge.
This entry is adapted from the peer-reviewed paper 10.3390/polym15081850
References
- Pal, S.; Tatini, R. Folate-Functionalized DNA Origami for Targeted Delivery of Doxorubicin to Triple-Negative Breast Cancer. Front. Chem. 2021, 9.
- Seeman, N.C.; Sleiman, H.F. DNA Nanotechnology. Nat. Rev. Mater. 2017, 3, 17068.
- Machtakova, M.; Thérien-Aubin, H.; Landfester, K. Polymer nano-systems for the encapsulation and delivery of active biomacromolecular therapeutic agents. Chem. Soc. Rev. 2022, 51, 128–152.
- Palaria, B.; Tiwari, V.; Tiwari, A.; Aslam, R.; Kumar, A.; Sahoo, B.M.; Kumar, M.; Singh, S.; Kumar, S. Nanostructured Lipid Carriers: A Promising Carrier in Targeted Drug Delivery System. Curr. Nanomat. 2023, 8, 23–43.
- Khatir, N.M.; Abdul-Malek, Z.; Banihashemian, S.M. Influences of magnetic fields on current–voltage characteristics of gold-DNA-gold structure with variable gaps. Mater. Sci. Semicond. Process. 2015, 36, 134–139.
- Khatir, N.M.; Sabbagh, F. Green Facile Synthesis of Silver-Doped Zinc Oxide Nanoparticles and Evaluation of Their Effect on Drug Release. Materials 2022, 15, 5536.
- Khatir, N.M.; Abdul-Malek, Z.; Zak, A.K.; Akbari, A.; Sabbagh, F. Sol–gel grown Fe-doped ZnO nanoparticles: Antibacterial and structural behaviors. J. Sol-Gel Sci. Technol. 2016, 78, 91–98.
- Weiden, J.; Bastings, M.M. DNA origami nanostructures for controlled therapeutic drug delivery. Curr. Opin. Colloid Interface Sci. 2021, 52, 101411.
- Schneider, F.; Möritz, N.; Dietz, H. The sequence of events during folding of a DNA origami. Sci Adv. 2019, 5.
- Andersen, E.S.; Dong, M.; Nielsen, M.M. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano 2008, 2, 1213–1218.
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 2009, 459, 414–418.
- Jun, H.; Wang, X.; Bricker, W.P.; Bathe, M. Automated sequence design of 2D wireframe DNA origami with honeycomb edges. Nat. Commun. 2019, 10, 5419.
- Benson, E.; Mohammed, A.; Gardell, J. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444.
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into twisted and curved nanoscale shapes. Science 2009, 325, 725–730.
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA origami with complex curvatures in three-dimensional space. Science 2011, 332, 342–346.
- Wagenbauer, K.F.; Sigl, C.; Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 2017, 552, 78–83.
- Han, D.; Qi, X.; Myhrvold, C. Single-stranded DNA and RNA origami. Science 2017, 358, 83–89.
- Gerling, T.; Wagenbauer, K.F.; Neuner, A.M.; Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 2015, 347, 1446–1452.
- Ge, Z.; Gu, H.; Li, Q.; Fan, C. Concept and Development of Framework Nucleic Acids. J. Am. Chem. Soc. 2018, 140, 17808–17819.
- Liu, X.; Zhang, F.; Jing, X. Complex silica composite nanomaterials templated with DNA origami. Nature 2018, 559, 593–598.
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.; Saccà, B.; Simmel, F.C.; et al. DNA origami. Nat. Rev. Met. Prim. 2021, 1, 13.
- Liu, N.; Liedl, T. DNA-Assembled Advanced Plasmonic Architectures. Chem. Rev. 2018, 118, 3032–3053.
- Yurke, B.; Turberfield, A.J.; Mills, A.P.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608.
- Li, S.; Jiang, Q.; Liu, S. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264.
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302.
- Nangreave, J.; Han, D.; Liu, Y.; Yan, H. DNA origami: A history and current perspective. Curr. Opin. Chem. Biol. 2010, 14, 608–615.
- Zhang, Q.; Jiang, Q.; Li, N. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 2014, 8, 6633–6643.
- Baig, M.M.F.A.; Xia, X.H. The PA-receptor mediated internalization of carboplatin loaded poly-anionic DNA-nanowires for effective treatment of resistant hepatic-cancer HepG-2 cells. Appl. Nanosci. 2020, 10, 1915–1926.
- Mariconti, M. DNA-Protein Nanogels as a New Class of Tunable Nanobiomaterials: From Enzymatic Nanoreactors to Transfection of Active Proteins. Ph.D. Thesis, Université Paris sciences et Lettres, Paris, France, 2021.
- Wang, D.X.; Wang, J.; Wang, Y.X.; Du, Y.C.; Huang, Y.; Tang, A.N.; Cui, Y.X.; Kong, D.M. DNA nanostructure-based nucleic acid probes: Construction and biological applications. Chem. Sci. 2021, 12, 7602–7622.
- Adamczyk, A.K.; Huijben, T.A.; Sison, M.; Di Luca, A.; Chiarelli, G.; Vanni, S.; Brasselet, S.; Mortensen, K.I.; Stefani, F.D.; Pilo-Pais, M.; et al. DNA self-assembly of single molecules with deterministic position and orientation. ACS Nano 2022, 16, 16924–16931.
- Ijäs, H.; Shen, B.; Heuer-Jungemann, A.; Keller, A.; Kostiainen, M.A.; Liedl, T.; Ihalainen, J.A.; Linko, V. Unraveling the interaction between doxorubicin and DNA origami nanostructures for customizable chemotherapeutic drug release. Nucleic Acids Res. 2021, 49, 3048–3062.
- Halley, P.D.; Lucas, C.R.; McWilliams, E.M. Daunorubicin-Loaded DNA Origami Nanostructures Circumvent Drug-Resistance Mechanisms in a Leukemia Model. Small 2016, 12, 308–320.
- Zhuang, X.; Ma, X.; Xue, X. A Photosensitizer-Loaded DNA Origami Nanosystem for Photodynamic Therapy. ACS Nano 2016, 10, 3486–3495.
- Wu, T.; Liu, J.; Liu, M. A Nanobody-Conjugated DNA Nanoplatform for Targeted Platinum-Drug Delivery. Angew Chem. Int. Ed. Engl. 2019, 58, 14224–14228.
- Nussbaumer, S.; Bonnabry, P.; Veuthey, J.L.; Fleury Souverain, S. Analysis of anticancer drugs: A review. Talanta 2011, 85, 2265–2289.
- Thurston, D.E.; Pysz, I. Chemistry and Pharmacology of Anticancer Drugs; CRC Press: Boca Raton, FL, USA, 2021; Volume 89, pp. 323–330.
- Bu, Y.Z.; Xu, J.R.; Luo, Q.; Chen, M.; Mu, L.M.; Lu, W.L. A precise nanostructure of folate-overhung mitoxantrone dna tetrahedron for targeted capture leukemia. Nanomaterials 2020, 10, 951.
- Sala, L.; Perecko, T.; Mestek, O.; Pinkas, D.; Homola, T.; Kocisek, J. Cisplatin-Cross-Linked DNA Origami Nanostructures for Drug Delivery Applications. ACS Appl. Nano Mater. 2022, 5, 13267–13275.
- Nathiya, S.; Durga, M.; Thiyagarajan, D. Quercetin, encapsulated quercetin and its application—A review. Int. J. Pharm. Pharm. Sci. 2014, 32, 20–26.
- Lin, Y.; Shi, R.; Wang, X.; Shen, H.M. Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr. Cancer Drug Targets 2008, 8, 634–646.
- Koba, M.; Konopa, J. Actinomycin D and its mechanisms of action. Postepy Hig. Med. Dosw. 2005, 59, 290–298.
- Mei, Q.; Wei, X.; Su, F. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 2011, 11, 1477–1482.
- Kearney, C.J.; Lucas, C.R.; O’Brien, F.J.; Castro, C.E. DNA origami: Folded DNA-nanodevices that can direct and interpret cell behavior. Adv. Mater. 2016, 28, 5509–5524.
- Yu, C.; An, M.; Jones, E.; Liu, H. Targeting Suppressive Oligonucleotide to Lymph Nodes Inhibits Toll-like Recep-tor-9-Mediated Activation of Adaptive Immunity. Pharm. Res. 2018, 35, 56–58.
- Li, J.; Pei, H.; Zhu, B. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano 2011, 5, 8783–8789.
- Huang, E.; Showalter, L.; Xu, S.; Czernliecki, B.J.; Koski, G.K. Calcium mobilizing treatment acts as a co-signal for TLR-mediated induction of Interleukin-12 (IL-12p70) secretion by murine bone marrow-derived dendritic cells. Cell Immunol. 2017, 314, 26–35.
- Chi, Q.; Yang, Z.; Xu, K.; Wang, C.; Liang, H. DNA nanostructure as an efficient drug delivery platform for immunotherapy. Front. Pharmacol. 2020, 10, 1585–1589.
- Xu, T.; Yu, S.; Sun, Y. DNA Origami Frameworks Enabled Self-Protective siRNA Delivery for Dual Enhancement of Chemo-Photothermal Combination Therapy. Small 2021, 17, 210–215.
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834.
- Jiang, Q.; Shi, Y.; Zhang, Q. A Self-Assembled DNA Origami-Gold Nanorod Complex for Cancer Theranostics. Small 2015, 11, 5134–5141.
- Du, Y.; Jiang, Q.; Beziere, N. DNA-Nanostructure-Gold-Nanorod Hybrids for Enhanced In Vivo Optoacoustic Imaging and Photothermal Therapy. Adv. Mater. 2016, 28, 10000–10007.
- Pan, M.; Jiang, Q.; Sun, J. Programming DNA Nanoassembly for Enhanced Photodynamic Therapy. Angew Chem. Int. Ed. Engl. 2020, 59, 1897–1905.
- Jiang, Q.; Liu, S.; Liu, J. Rationally designed DNA-origami nanomaterials for drug delivery in vivo. Adv. Mater. 2018, 31, 1804785.
- Elbaz, J.; Yin, P.; Voigt, C.A. Genetic encoding of DNA nanostructures and their self-assembly in living bacteria. Nat. Commun. 2016, 7, 11–17.
- Veneziano, R.; Ratanalert, S.; Zhang, K. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 15–34.
- Hahn, J.; Wickham, S.F.; Shih, W.M.; Perrault, S.D. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 2014, 8, 8765–8775.
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The beauty and utility of DNA origami. Chem 2017, 2, 359–382.
- Li, J.; Fan, C.; Pei, H.; Shi, J.; Huang, Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv. Mater. 2013, 25, 4386–4396.
- Schüller, V.J.; Heidegger, S.; Sandholzer, N. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano 2011, 5, 9696–9702.
- Zhao, Z.; Fu, J.; Dhakal, S. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 2016, 7, 10619.
- Yan, J.; Hu, C.; Wang, P. Growth and origami folding of DNA on nanoparticles for high-efficiency molecular transport in cellular imaging and drug delivery. Angew Chem. Int. Ed. Engl. 2015, 54, 2431–2435.
- Kong, F.; Zhang, H.; Qu, X.; Zhang, X.; Chen, D.; Ding, R. Gold nanorods, DNA origami, and porous silicon nanoparticle-functionalized biocompatible double emulsion for versatile targeted therapeutics and antibody combination therapy. Adv. Mater. 2016, 28, 10195–10203.
- Oleinick, N.L.; Morris, R.L.; Belichenko, I. The role of apoptosis in response to photodynamic therapy: What, where, why, and how. Photochem. Photobiol. Sci. 2002, 1, 1–21.
- Jiang, Q.; Song, C.; Nangreave, J.; Liu, X.; Lin, L.; Qiu, D. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403.
