Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Immunology
The CC chemokine receptor 6 (CCR6) is a G protein-coupled receptor (GPCR) involved in a wide range of biological processes. When CCR6 binds to its sole ligand CCL20, a signaling network is produced. This pathway is implicated in mechanisms related to many diseases, such as cancer, psoriasis, multiple sclerosis, HIV infection or rheumatoid arthritis.
- CCR6
- antibody
- therapy
1. Introduction
G protein-coupled receptors (GPCRs) are one of the most abundant receptors encoded in the human genome, with over 800 members, and transmit approximately 80% of signal transduction across cell membranes [1,2]. GPCRs signal through activation of Gα, Gβ and Gγ subunits of heterotrimeric G protein and β-arrestin protein mediators [3]. GPCRs are involved in a broad range of key biological processes, including homeostasis, proliferation and chemotaxis of cells, and have been implicated in a considerable number of diseases, such as cancer, inflammation and infection [4,5].
Chemokine receptors are a family of GPCRs regulated by small ligands known as chemokines. These molecules are low molecular weight proteins with a globular core structure stabilized by 1–2 conserved disulfide bridges essential in leukocyte trafficking through the formation of chemotactic gradients [6,7,8]. Chemokines and chemokine receptors play important roles in a broad range of biological and pathological processes controlling the activation, migration, differentiation and survival of leukocytes and other hematopoietic cells [9,10].
The CC chemokine receptor 6 (CCR6) is a class A GPCR belonging to the chemokine family and is recognized for its invaluable therapeutic potential in research related to the immune system [11]. CCR6 is expressed in numerous cells, including B cells, immature dendritic cells (DCs), innate lymphoid cells (ILCs), Langerhans cells, neutrophils, regulatory T (Treg) cells and T helper 17 (Th17) cells [12,13]. The only chemokine ligand of CCR6 is CCL20 and, in humans and mice, is expressed by neutrophils, Th17 cells and peripheral blood mononuclear cells [8,14,15]. This axis plays exclusive roles in immune homeostasis and activation. The influence of the CCR6/CCL20 partnership via a pleiotropic immune mechanism in the respiratory, nervous, excretory, skeletal, gastrointestinal and reproductive systems has been demonstrated, manifesting as numerous diseases [11].
Although the relationship between CCR6 and many diseases has been widely studied, at this moment, there is no therapeutic agent against CCR6 approved [16]. Given the important roles that CCR6 and CCL20 play in clinical pathophysiology, this axis is considered a potential therapeutic target. An antagonizing monoclonal antibody could be a potential alternative to conventional small-molecule drugs and an effective strategy for treating certain inflammatory and autoimmune diseases.
2. Description of CCR6
2.1. Biochemical Characteristics and Structure
The human CCR6 gene was mapped to chromosome 6q27, outside the main cluster of CC chemokine receptor genes on chromosome 3p [17]. The CCR6 receptor is a protein embedded in the cell membrane with seven transmembrane α-helices connected by three extracellular loops (ECL1–3) and three intracellular loops (ICL1–3). The extracellular (EC) region, which is responsible for ligand binding, also contains the N-terminus and the intracellular (IC) region, which includes cytoplasmic helix H8 and a C-terminus, interacts with G proteins, arrestins and other downstream effectors [18].
The only known high-affinity chemokine ligand of the CCR6 receptor is CCL20; however, low-affinity binding of the human beta-defensins (hBDs)-1 and -2, a group of anti-bacterial peptides, to CCR6, has been reported [6]. Some chemokines bind to more than one receptor, but CCL20 binds specifically to the CCR6 receptor and forms an exclusive, monogamous pair [19]. The binding site of CCL20 to CCR6 is a shallow extracellular pocket, in contrast to the deep agonist-binding sites observed in other class A GPCRs, producing interactions with the three ECLs. Moreover, the N-terminal residues Y27 and L38 from the receptor wrap onto the globular core of CCL20, serving as another critical docking of chemokine binding [8].
CCR6 is composed of 139,737 bases long and encodes a protein with 374 amino acids with a molecular weight of 42 kDa [20]. Wasilko et al. elucidated a cryo-electron microscopy (Cryo-EM) structure of human CCR6 receptor bound to CCL20 and Go protein (Figure 1), giving important insights into the mechanism of activation of CCR6 [8]. Furthermore, a homology model of human CCR6 is available in the GPCR database (GPCRdb). Snake and helix box diagrams are 2D receptor topology accessible plots that map the position of binding residues as seen from the extracellular and membrane sides, respectively [21].
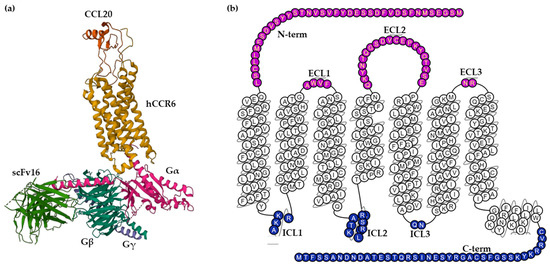
Figure 1. (a) Cryo-EM structure of the human chemokine receptor CCR6 in complex with CCL20 and a Go protein (PDB ID: 6WWZ) colored by subunit. Wasilko et al. introduced the single-chain variable fragment (scFv16) in the complex to slow down the dissociation. (b) Snake helix box diagram depicts human CCR6 topology as seen from the side (GPCR database). Extracellular domains: purple; transmembrane domains: white; intracellular domains: blue.
2.2. Expression of CCR6
Human tissue expression of CCR6 can be predominantly seen in the appendix, pancreas, lymph nodes, spleen and, with lesser expression, in the fetal liver, testis, colon, small intestine and thymus [13,22].
At the cellular level, CCR6 is expressed in a variety of immune cell types (Table 1), consistent with its well-established role in inflammation. There are numerous leukocyte cohorts such as B cells, dendritic cells, innate lymphoid cells 3 (ILC-3), T cells, specifically pro-inflammatory Th17 cells and immune regulatory Treg cells, and neutrophils in which CCR6 is upregulated [19,22,23,24,25,26,27,28,29,30].
Table 1. CCR6+ cells.
| Cell Type | Reference |
|---|---|
| B cell | [19,22,23,24] |
| Immature DC | [19,22,24,25] |
| ICL-3 | [19,22,25] |
| Langerhans cell | [26,27] |
| NK cell | [17,28] |
| NKT cell | [19,22,26] |
| Neutrophils | [19,22] |
| Th9 cell | [28] |
| Th17 cell | [19,22,23,29] |
| Th22 cell | [28,30] |
| Treg cell | [19,22,23,24] |
| γδT cell | [28] |
DC—dendritic cell; ILC-3—innate lymphoid cell 3; NK cell—natural killer cell; NKT cell—natural killer T cell; Th cell—T helper cell; Treg cell—regulatory T cell.
CCR6 is found in various B cell subtypes [13]. The receptor is expressed at the cell surface of circulating, naïve and memory but not germinal center B cells. On neutrophils, it has been reported that CCR6 can be upregulated after treatment with cytokines in vitro [24]. ILC-3 cells and immature DCs also expressed CCR6, although its expression on immature DC is lost following their maturation [25]. Moreover is known that CCR6 is expressed on multiple DCs subsets, including Langerhans cells [26].
Natural killer T (NKT) cells are a T cell subset that expresses natural killer (NK) cell markers and represent about 0.1% of peripheral blood lymphocytes. Both cells were shown to express CCR6 and be attracted by CCL20 [17]. CCR6 was found to be more frequently present in CD4+ T cells than in CD8+ T cells [17]. The CCR6 receptor plays a pivotal role in the regulation of T cells to inflammatory sites, and these cells are recruited by CCL20 interaction with CCR6 [28].
Quantitative information provided by The Human Protein Atlas of RNA expression across single cell types confirms the expression of CCR6 in cells shown in Table 1. The normalized single-cell RNA (nTPM) values for CCR6 are 13.5 in T cells, 15.9 in B cells, 1.2 in plasma cells, 1.4 in NK cells, 0.1 in monocytes, 0.4 in macrophages, 0.4 in dendritic cells and 0.8 in Langerhans cells [31]. The reciprocal data of normalized single-cell RNA (nTPM) for CCL20 are 46.6 in T cells, 2.1 in B cells, 3.3 in plasma cells, 1.9 in NK cells, 1275.2 in monocytes, 214.7 in macrophages, 15.2 in dendritic cells and 210.7 in Langerhans cells [32]. The data indicates a high correlation between the expression of CCR6 and CCL20 in T cells.
2.3. Signaling Pathways of CCR6
Activation of CCR6 elicits a combination of responses, including activation of G proteins and β-arrestins mediated signal transduction, and both have different non-overlapping functions and play important roles in CCR6 signaling [33].
When G proteins are activated, they dissociate from the receptor, and their α- and βγ-subunits separate and trigger the activation of a second messenger signaling and multiple intracellular pathways [34]. Upon binding the ligand to CCR6, the receptor signaling is predominantly via Gαi proteins which inhibit the cyclic 3′,5′-cyclic monophosphate (cAMP)-dependent pathway through the inhibition of adenylate cyclase [8]. Additionally, activation of CCR6 induces the release of the Gβγ subunit from the Gα subunit, which activates phospholipase C (PLC) and triggers an increase in intracellular Ca2+ levels [35]. When CCL20 binds to CCR6, it initiates the activation of calcium mobilization, phospholipase and phophatidyinositol-3-kinase, followed by ERK1/2 phosphorylation and actin polymerization [19].
Furthermore, like most chemokine receptors, CCR6 can interact with additional effectors such as β-arrestin. This activation involves CCR6 phosphorylation by GPCR kinases (GRK) and the β-arrestin recruitment causing receptor desensitization and internalization [7,36]. The complex CCR6–arrestin acts as a scaffold facilitating different signaling pathways through c-Src, ERK 1/2, p38, JNK or Akt, initiating a G protein-independent wave of downstream signaling [36,37,38].
CCR6 can be activated by the union of the high-affinity ligand, the CCL20 chemokine, or human β-defensins that have been reported as non-chemokine low-affinity ligands of CCR6 [14,17]. The ligand binds to the receptor recognition site formed by the turn between β2- and β3-strands and the N-terminal loop. The N-terminus of CCL20 makes key contacts with the transmembrane bundle of CCR6 for stimulating intracellular signaling and receptor activation [39,40]. The signaling by the CCR6/CCL20 axis is critical for humoral immune responses and plays an important role in the migration of Th17 cells to inflamed sites. The signaling of CCR6 by hBDs interaction is able to induce chemotaxis in immature dendritic cells and memory T cells, but its activity remains to be investigated [41,42].
This entry is adapted from the peer-reviewed paper 10.3390/antib12020030
This entry is offline, you can click here to edit this entry!
