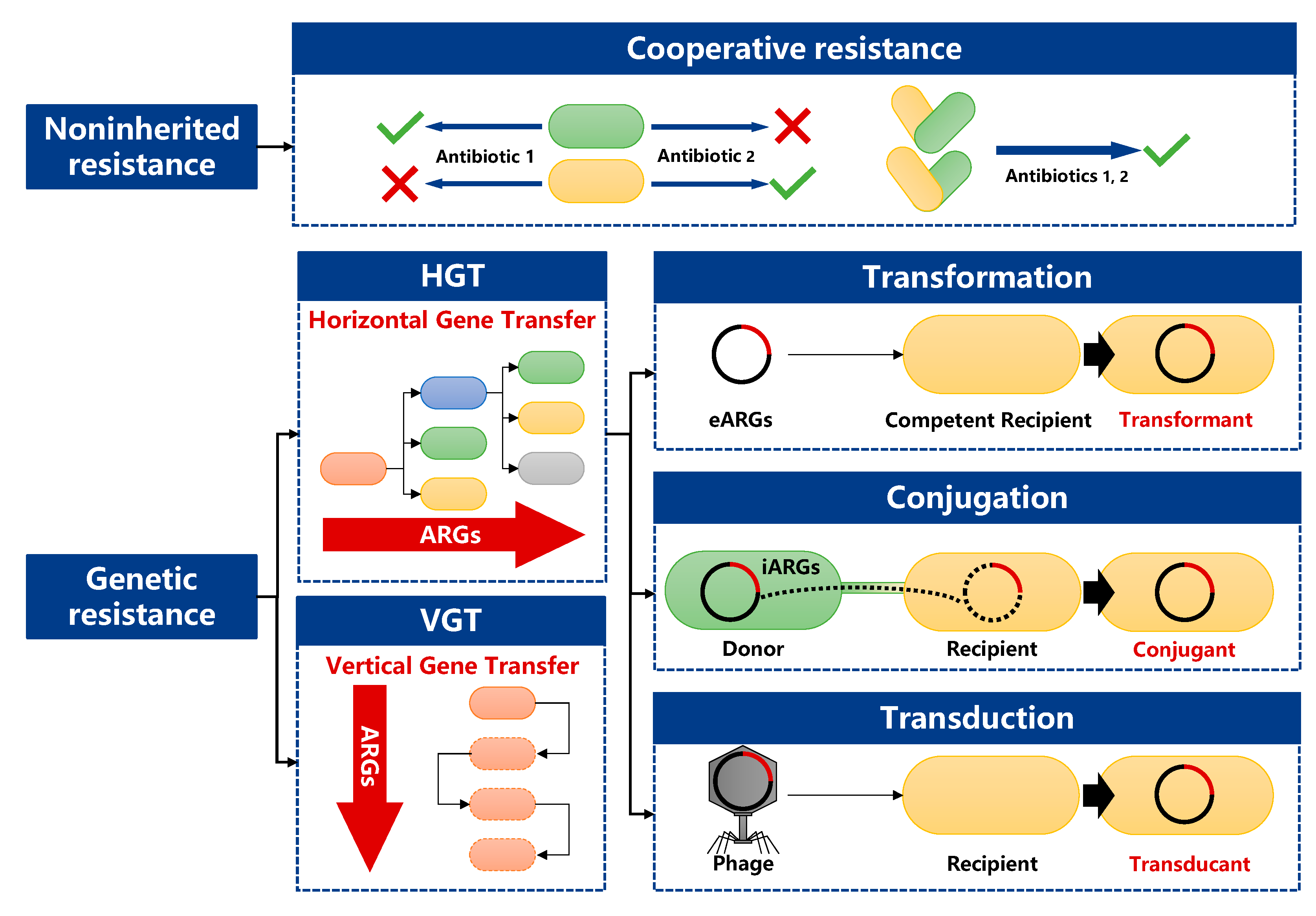
Antibiotic resistance genes (ARGs) and antibiotic-resistant bacteria (ARB) in soil have become research hotspots in the fields of public health and environmental ecosystem. The soil environment is an important acceptor of many pollutants, including antibiotics. The external pressure of antibiotics and other pollutants can promote the proliferation and occurrence of antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARGs) in soil. Soil ARGs can spread in various ways, such as horizontal gene transfer (HGT) between soil microorganisms and vertical gene transfer (VGT) between parent and offspring, while soil components have important influence on the occurance and spread of ARGs.
- soil component
- antibiotic resistance genes
- horizontal gene transfer
1. The Occurrence and Spread of ARGs in Soil
1.1. The Pollution Status of ARB and ARGs in Soil
| Place | Soil Type | ARGs | Relative Abundance |
|---|---|---|---|
| China [13] | Feedlot vicinity | tetM, tetO, tetQ, tetW | 10−5–10−2 |
| China [14] | Feedlot vicinity | tetB(P), tetM, tetO, tetW | 10−3–10−6 |
| China [15] | Feedlots | tetA(P), tetG, tetC, tetL, tetX, tetM, tetA | 10−2–10−4 |
| China [16] | Feedlots | tetA, tetB, tetM | 10−6–10−1 |
| China [17] | Farmland | tetB(P), tetM, tetO, tetQ, tetT, tetW | 10−8–10−2 |
| China [18] | Farmland | tetG, tetY, tetZ | 10−7–10−4 |
| China [19] | Farmland | tetB(P), tetC, tetG, tetL, tetO, tetS, tetW, tetZ | 10−6–10−1 |
| Italy [20] | Feedlots | tetQ, tetW | 10−9–10−5 |
| India [21] | Feedlots | tetA, tetW | 10−1 a |
| America [22] | Farmland | tetO, tetW | 10−7–10−4 |
| Austria [23] | Farmland | tetW | 10−5–10−4 |
| The Netherlands [11] | Typical sites | tetM, tetO, tetQ, tetW | 10−4–10−2 |
| Scotland [3] | Typical sites | tetM, tetQ, tetW | 10−5–10−2 |
| Scotland [24] | Farm | tetA, tetB, tetC, tetG, tetW | 10−6–10−5 |
| Australia [25] | Residential area | tetM, tetW | 10−9–10−2 |
1.2. Transmission Routes of ARGs
2. Effects of Soil Components on the HGT of ARGs from the Micro Perspective
2.1. The Effects of Soil Components on the HGT of ARGs in Pure Bacterial System
| Medium | Important Conclusions | |
|---|---|---|
| Results | Reasons | |
| Kaolinite, illite, and montmorillonite [69] | Plasmids adsorbed on minerals could resist higher concentrations of nucleases and form more transformants than free plasmids. | The adsorption of the nuclease on minerals protected the plasmids, but it can still be involved in transformation. |
| Kaolinite, Goethite, and montmorillonite [70] |
Low concentrations (1–2 g/L) have little effect; high concentration (10 g/L) of kaolinite and montmorillonite inhibited transformation; high concentration (10 g/L) of goethite promoted transformation. | Kaolinite and montmorillonite: strong adsorption to competence stimulating factor, decrease the expression level of competent genes (phrC, comS); goethite: increase cell membrane damage. |
| Montmorillonite [71] | Low concentration (about 0–0.025 g/L) promoted transformation; high concentration (about 0.025–2 g/L) inhibited transformation. |
Low concentration: increase the contact between plasmids and cells; forming holes on cell membrane; High concentration: plasmids were adsorbed; heavy metals released from montmorillonite cause the aggregation of the plasmids. |
| Biochar [72] | Significantly inhibited the transformation of extracellular antibiotic resistance genes (eARGs) | Biochar dissolutions: Induce intramolecular condensation and agglomeration of plasmids; decrease the cell membrane permeability; biochar solids: Adsorb plasmids and deactivate E. coli. |
| Soil microcosm [73] | DNA adsorbed on soil particles still transformed competent cells | Minerals did not inhibit the transformation, but blocked DNA contact with the recipient. |
| Soil microcosm [74] | Plasmid adsorbed on sand transformed significantly less efficient than did plasmid in solution; the transformation by sand-adsorbed chromosomal was as high as that by plasmid in solution. |
Transformation occurred by direct uptake of DNA from the mineral surfaces; transformation requires multiple plasmids, and the probability of multiple free plasmids meeting bacteria at the same time is higher than that on mineral surfaces; the chances of bacteria taking up DNA on the mineral surface are proportional to the size of the DNA, and chromosomes of the same mass are larger and easier to take up. |
| Activated sludge EPS [75] | The transformation ability of free ARGs was higher than that in activated sludge extracellular polymeric substances (EPS) when calculated per ng DNA, and lower when calculated per g volatile suspended solids. | Activated sludge EPS is rich in ARGs. |
| Sediment [76] | The transformation efficiency of adsorbed eARG was higher than that of free eARGs. | Sand adsorbed bacteria and plasmids at the same time, facilitating contact between the two, and was related to the conformation of the plasmid. |
| Medium | Results | Reasons |
|---|---|---|
| Kaolinite, goethite, birnessite, and montmorillonite [77] | Birnessite promoted conjugation. The effects of kaolinite and montmorillonite were irregular. Goethite promoted conjugation at low concentration (0–0.5 g/L) and inhibited it at high concentration (5 g/L). |
Birnessite promoted the production of intracellular reactive oxygen species (ROS); increased the expression levels of oxidative stress-regulated genes (rpoS) and outer membrane protein genes (ompA, ompF, ompC). Birnessite altered the expression levels of conjugation-related genes (globally regulation genes (korA, korB, trbA); mating pair formation (MPF) system genes (trbBp, traF); DNA transfer and replication (DTR) system genes (trfAp, traJ)). |
| Dissolved biochar [81] | The effects on conjugation were related to the concentration and source of biochar. | Humic acid-like substance in dissolved biochar improved the conjugative efficiency. The inhibitory effects of small-molecule matters dominated, decreasing conjugative transfer frequency. |
| Pyroligneous acid and its three fractions [79] | Reduced the abundance of ARGs and MGEs in soil. | High content of organic acids inhibited the bacterial growth. |
| Dissolved biochar [78] | Attenuated the promotion effect of Cu (Ⅱ) to conjugation. | Dissolved biochar affected intracellular ROS production level, cell membrane permeability, and the expression level of global regulatory genes (korA, korB, trbA), pore formation and membrane trafficking genes (ompA, ompC), MPF system gene (trbB), DTR system gene (trfA), etc. |
| CeO2 nanoparticle [80] (soil pollutant) |
Inhibited conjugation at low concentration (1, 5 mg/L), while promoted it at high concentration (25, 50 mg/L). | CeO2 nanoparticle affected many aspects, such as intracellular ROS production, polysaccharide synthesis in EPS, cell-to-cell contact, ATP supply, and the expression level of conjugation-related genes (MPF system gene (trbBp), DTR system gene (trfAp), putative transmembrane ATPase gene (traG)), etc. |
| Gut of C. elegans [82] (soil animal) |
The conjugation efficiency in gut was higher than soil, and increased with time and temperature. | The abundance of MPF system gene (trbBp) and DTR system gene (trfAp) was increased. |
2.2. Influence Mechanisms of Soil Components on HGT of ARGs
2.2.1. Intracellular Changes and Responses
Intracellular ROS Production
SOS Response
Cell Membrane Permeability
ATP Synthesis Capacity
Conjugation Activity of Intracellular Plasmids
2.2.2. Cell-Cell Contact and Quorum Sensing
2.2.3. Bacterial Uptake of Extracellular ARGs
The Competent State of Bacteria
Availability of Extracellular ARGs
2.2.4. Bacterial Concentration
This entry is adapted from the peer-reviewed paper 10.3390/antibiotics12020333
References
- Nesme, J.; Simonet, P. The soil resistome: A critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ. Microbiol. 2015, 17, 913–930.
- Ondon, B.S.; Li, S.N.; Zhou, Q.X.; Li, F. Sources of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in the soil: A review of the spreading mechanism and human health risks. In Reviews of Environmental Contamination and Toxicology; de Voogt, P., Ed.; Springer International Publishing: Cham, Switzerland, 2021; Volume 256, pp. 121–153.
- Knapp, C.W.; McCluskey, S.M.; Singh, B.K.; Campbell, C.D.; Hudson, G.; Graham, D.W. Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS ONE 2011, 6, e27300.
- Knapp, C.W.; Zhang, W.; Sturm, B.S.M.; Graham, D.W. Differential fate of erythromycin and beta-lactam resistance genes from swine lagoon waste under different aquatic conditions. Environ. Pollut. 2010, 158, 1506–1512.
- He, J.Z.; Yan, Z.Z.; Chen, Q.L. Transmission of antibiotic resistance genes in agroecosystems: An overview. Front. Agric. Sci. Eng. 2020, 7, 329–332.
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461.
- Xu, H.; Chen, Z.Y.; Huang, R.Y.; Cui, Y.; Li, Q.; Zhao, Y.; Wang, X.; Mao, D.; Luo, Y.; Ren, H.Q. Antibiotic resistance gene-carrying plasmid spreads into the plant endophytic bacteria using soil bacteria as carriers. Environ. Sci. Technol. 2021, 55, 10462–10470.
- Liu, J.X.; Zhao, Z.; Avillan, J.J.; Call, D.R.; Davis, M.; Sischo, W.M.; Zhang, A. Dairy farm soil presents distinct microbiota and varied prevalence of antibiotic resistance across housing areas. Environ. Pollut. 2019, 254, 113058.
- Furlan, J.P.R.; Stehling, E.G. Multiple sequence types, virulence determinants and antimicrobial resistance genes in multidrug- and colistin-resistant Escherichia coli from agricultural and non-agricultural soils. Environ. Pollut. 2021, 288, 117804.
- Graves, A.K.; Liwimbi, L.; Israel, D.W.; van Heugten, E.; Robinson, B.; Cahoon, C.W.; Lubbers, J.F. Distribution of ten antibiotic resistance genes in E. coli isolates from swine manure, lagoon effluent and soil collected from a lagoon waste application field. Folia Microbiol. 2011, 56, 131–137.
- Knapp, C.W.; Dolfing, J.; Ehlert, P.A.I.; Graham, D.W. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 2010, 44, 580–587.
- Zhang, Y.; Hu, X.; Wang, X. Recent advances on antibiotic resistance genes encoded by bacteriophages. China Environ. Sci. 2022, 42, 2315–2320.
- Wu, N.; Qiao, M.; Zhang, B.; Cheng, W.-D.; Zhu, Y.-G. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ. Sci. Technol. 2010, 44, 6933–6939.
- Ji, X.L.; Shen, Q.H.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235, 178–185.
- Wang, F.H.; Qiao, M.; Chen, Z.; Su, J.-Q.; Zhu, Y.-G. Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J. Hazard. Mater. 2015, 299, 215–221.
- Liu, Z.B.; Klümper, U.; Shi, L.; Ye, L.; Li, M. From pig breeding environment to subsequently produced pork: Comparative analysis of antibiotic resistance genes and bacterial community composition. Front. Microbiol. 2019, 10, 43.
- Zhou, Y.; Niu, L.; Zhu, S.; Lu, H.; Liu, W. Occurrence, abundance, and distribution of sulfonamide and tetracycline resistance genes in agricultural soils across China. Sci. Total Environ. 2017, 599, 1977–1983.
- Liu, L.; Huang, W.; Lyu, X.; He, X.; Chen, L.; Song, Y. Effect of Long-term Biogas Slurry Fertilization on Distribution of Tetracycline and Sulfonamide Resistance Genes in Soil. Fujian J. Agric. Sci. 2021, 36, 699–705.
- Peng, S.; Wang, Y.; Lin, X. Abundance of the tetracycline resistance genes in a paddy soil after continuous application of composted swine manure for 6 years. China Environ. Sci. 2015, 35, 1173–1180.
- Chessa, L.; Jechalke, S.; Ding, G.C.; Pusino, A.; Mangia, N.P.; Smalla, K. The presence of tetracycline in cow manure changes the impact of repeated manure application on soil bacterial communities. Biol. Fertil. Soils 2016, 52, 1121–1134.
- Anand, T.; Bera, B.C.; Vaid, R.K.; Barua, S.; Riyesh, T.; Virmani, N.; Hussain, M.; Singh, R.K.; Tripathi, B.N. Abundance of antibiotic resistance genes in environmental bacteriophages. J. Gen. Virol. 2016, 97, 3458–3466.
- Munir, M.; Xagoraraki, I. Levels of antibiotic resistance genes in manure, biosolids, and fertilized soil. J. Environ. Qual. 2011, 40, 248–255.
- Radu, E.; Woegerbauer, M.; Rab, G.; Oismüller, M.; Strauss, P.; Hufnagl, P.; Gottsberger, R.A.; Krampe, J.; Weyermair, K.; Kreuzinger, N. Resilience of agricultural soils to antibiotic resistance genes introduced by agricultural management practices. Sci. Total Environ. 2021, 756, 143699.
- Lin, H.; Chapman, S.J.; Freitag, T.E.; Kyle, C.; Ma, J.; Yang, Y.; Zhang, Z. Fate of tetracycline and sulfonamide resistance genes in a grassland soil amended with different organic fertilizers. Ecotoxicol. Environ. Saf. 2019, 170, 39–46.
- Knapp, C.W.; Callan, A.C.; Aitken, B.; Shearn, R.; Koenders, A.; Hinwood, A. Relationship between antibiotic resistance genes and metals in residential soil samples from western Australia. Environ. Sci. Pollut. Res. 2017, 24, 2484–2494.
- Corona, F.; Martinez, J.L. Phenotypic Resistance to Antibiotics. Antibiotics 2013, 2, 237–255.
- Frost, I.; Smith, W.P.J.; Mitri, S.; San Millan, A.; Davit, Y.; Osborne, J.M.; Pitt-Francis, J.M.; MacLean, R.C.; Foster, K.R. Cooperation, competition and antibiotic resistance in bacterial colonies. ISME J. 2018, 12, 1582–1593.
- Sorg, R.A.; Lin, L.; van Doorn, G.S.; Sorg, M.; Olson, J.; Nizet, V.; Veening, J.-W. Collective resistance in microbial communities by intracellular antibiotic deactivation. PLoS Biol. 2016, 14, e2000631.
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433.
- Perron, G.G.; Whyte, L.; Turnbaugh, P.J.; Goordial, J.; Hanage, W.P.; Dantas, G.; Desai, M.M. Functional characterization of bacteria isolated from ancient Arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS ONE 2015, 10, e0069533.
- Pontes, D.S.; Aquino de Araujo, R.S.; Dantas, N.; Scotti, L.; Scotti, M.T.; de Moura, R.O.; Bezerra Mendonca-Junior, F.J. Genetic mechanisms of antibiotic resistance and the role of antibiotic adjuvants. Curr. Top. Med. Chem. 2018, 18, 42–74.
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173.
- Seoane, J.; Yankelevich, T.; Dechesne, A.; Merkey, B.; Sternberg, C.; Smets, B.F. An individual-based approach to explain plasmid invasion in bacterial populations. FEMS Microbiol. Ecol. 2011, 75, 17–27.
- Meng, M.; Li, Y.; Yao, H. Plasmid-mediated transfer of antibiotic resistance genes in soil. Antibiotics 2022, 11, 525.
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid transfer by conjugation in gram-negative bacteria: From the cellular to the community Level. Genes 2020, 11, 1239.
- Chen, M.-l.; An, X.-l.; Yang, K.; Zhu, Y.-g. Soil phage and their mediation on the horizontal transfer of antibiotic resistance genes: A review. Yingyong Shengtai Xuebao 2021, 32, 2267–2274.
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 2020, 96, fiaa031.
- Getino, M.; Cruz, F.D.L.; Baquero, F.; Bouza, E.; Gutiérrez-Fuentes, J.A.; Coque, T.M. Natural and artificial strategies to control the conjugative transmission of plasmids. Microbiol. Spectr. 2018, 6, MTBP-0015-2016.
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721.
- de Vries, J.; Wackernagel, W. Microbial horizontal gene transfer and the DNA release from transgenic crop plants. Plant Soil 2004, 266, 91–104.
- Li, S.N.; Zhang, C.F.; Li, F.X.; Hua, T.; Zhou, Q.X.; Ho, S.H. Technologies towards antibiotic resistance genes (ARGs) removal from aquatic environment: A critical review. J. Hazard. Mater. 2021, 411, 125148.
- Chen, P.; Chen, C.; Li, X. Transport of antibiotic resistance plasmids in porous media and the influence of surfactants. Front. Environ. Sci. Eng. 2018, 12, 5.
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304.
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-00017.
- Smillie, C.; Pilar Garcillan-Barcia, M.; Victoria Francia, M.; Rocha, E.P.C.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452.
- Waters, V.L. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 1999, 4, 433–456.
- Norman, A.; Hansen, L.H.; Sorensen, S.J. Conjugative plasmids: Vessels of the communal gene pool. Philos. Trans. R. Soc. B-Biol. Sci. 2009, 364, 2275–2289.
- Dionisio, F.; Conceicao, I.C.; Marques, A.C.R.; Fernandes, L.; Gordo, I. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 2005, 1, 250–252.
- Hall, J.P.J.; Williams, D.; Paterson, S.; Harrison, E.; Brockhurst, M.A. Positive selection inhibits gene mobilization and transfer in soil bacterial communities. Nat. Ecol. Evol. 2017, 1, 1348–1353.
- Clark, A.J.; Adelberg, E.A. Bacterial conjugation. Annu. Rev. Microbiol. 1962, 16, 289–319.
- Llosa, M.; Gomis-Ruth, F.X.; Coll, M.; de la Cruz, F. Bacterial conjugation: A two-step mechanism for DNA transport. Plasmid 2002, 48, 243.
- Fox, R.E.; Zhong, X.; Krone, S.M.; Top, E.M. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2008, 2, 1024–1039.
- Nielsen, K.M.; van Elsas, J.D. Stimulatory effects of compounds present in the rhizosphere on natural transformation of Acinetobacter sp BD413 in soil. Soil Biol. Biochem. 2001, 33, 345–357.
- Klumper, U.; Dechesne, A.; Riber, L.; Brandt, K.K.; Gulay, A.; Sorensen, S.J.; Smets, B.F. Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. Isme J. 2017, 11, 152–165.
- Fan, X.-T.; Li, H.; Chen, Q.-L.; Zhang, Y.-S.; Ye, J.; Zhu, Y.-G.; Su, J.-Q. Fate of Antibiotic Resistant Pseudomonas putida and Broad Host Range Plasmid in Natural Soil Microcosms. Front. Microbiol. 2019, 10, 194.
- Colombi, E.; Straub, C.; Kuenzel, S.; Templeton, M.D.; McCann, H.C.; Rainey, P.B. Evolution of copper resistance in the kiwifruit pathogen Pseudomonas syringae pv. actinidiae through acquisition of integrative conjugative elements and plasmids. Environ. Microbiol. 2017, 19, 819–832.
- Goncalves, O.S.; Santana, M.F. The coexistence of monopartite integrative and conjugative elements in the genomes of Acidobacteria. Gene 2021, 777, 145476.
- Furuya, E.Y.; Lowy, F.D. Antimicrobial-resistant bacteria in the community setting. Nat. Rev. Microbiol. 2006, 4, 36–45.
- Muniesa, M.; Colomer-Lluch, M.; Jofre, J. Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol. 2013, 8, 739–751.
- Pantasticocaldas, M.; Duncan, K.E.; Istock, C.A.; Bell, J.A. Population-Dynamics of Bacteriophage and Bacillus-Subtilis in Soil. Ecology 1992, 73, 1888–1902.
- Liao, H.; Li, H.; Duan, C.S.; Zhou, X.Y.; An, X.L.; Zhu, Y.G.; Su, J.Q. Metagenomic and viromic analysis reveal the anthropogenic impacts on the plasmid and phage borne transferable resistome in soil. Environ. Int. 2022, 170, 107595.
- Guemes, A.G.C.; Youle, M.; Cantu, V.A.; Felts, B.; Nulton, J.; Rohwer, F. Viruses as Winners in the Game of Life. Annu. Rev. Virol. 2016, 3, 197–214.
- Wilhelm, S.W.; Weinbauer, M.G.; Suttle, C.A.; Jeffrey, W.H. The role of sunlight in the removal and repair of viruses in the sea. Limnol. Oceanogr. 1998, 43, 586–592.
- Williamson, K.E.; Fuhrmann, J.J.; Wommack, K.E.; Radosevich, M. Viruses in soil ecosystems: An unknown quantity within an unexplored territory. Annu. Rev. Virol. 2017, 4, 201–219.
- Kenzaka, T.; Tani, K.; Sakotani, A.; Yamaguchi, N.; Nasu, M. High-frequency phage-mediated gene transfer among Escherichia coli cells, determined at the single-cell level. Appl. Environ. Microbiol. 2007, 73, 3291–3299.
- Subirats, J.; Sanchez-Melsio, A.; Borrego, C.M.; Luis Balcazar, J.; Simonet, P. Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes. Int. J. Antimicrob. Agents 2016, 48, 163–167.
- Larranaga, O.; Brown-Jaque, M.; Quiros, P.; Gomez-Gomez, C.; Blanch, A.R.; Rodriguez-Rubio, L.; Muniesa, M. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 2018, 115, 133–141.
- Li, B.; Qiu, Y.; Song, Y.Q.; Lin, H.; Yin, H. Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ. Int. 2019, 131, 105007.
- Demaneche, S.; Jocteur-Monrozier, L.; Quiquampoix, H.; Simonet, P. Evaluation of biological and physical protection against nuclease degradation of clay-bound plasmid DNA. Appl. Environ. Microbiol. 2001, 67, 293–299.
- Huang, Q.; Chen, J.X.; Zhu, J.J.; Hao, X.; Dao, G.; Chen, W.; Cai, P.; Huang, Q. Divergent bacterial transformation exerted by soil minerals. Sci. Total Environ. 2021, 784, 147173.
- Hu, X.J.; Sheng, X.; Zhang, W.; Lin, Z.; Gao, Y. Nonmonotonic effect of montmorillonites on the horizontal transfer of antibiotic resistance genes to bacteria. Environ. Sci. Technol. Lett. 2020, 7, 421–427.
- Fang, J.; Jin, L.; Meng, Q.K.; Shan, S.; Wang, D.; Lin, D.H. Biochar effectively inhibits the horizontal transfer of antibiotic resistance genes via transformation. J. Hazard. Mater. 2022, 423, 127150.
- Paget, E.; Simonet, P. Development of engineered genomic DNA to monitor the natural transformation of Pseudomonas stutzeri in soil-like microcosms. Can. J. Microbiol. 1997, 43, 78–84.
- Chamier, B.; Lorenz, M.G.; Wackernagel, W. Natural transformation of Acinetobacter calcoaceticus by plasmid DNA adsorbed on sand and groundwater aquifer material. Appl. Environ. Microbiol. 1993, 59, 1662–1667.
- Wang, L.; Yuan, L.; Li, Z.H.; Zhang, X.; Sheng, G.-P. Quantifying the occurrence and transformation potential of extracellular polymeric substances (EPS)-associated antibiotic resistance genes in activated sludge. J. Hazard. Mater. 2021, 408, 124428.
- Dong, P.Y.; Wang, H.; Fang, T.T.; Wang, Y.; Ye, Q.H. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96.
- Wu, S.; Wu, Y.C.; Huang, Q.Y.; Cai, P. Insights into conjugative transfer of antibiotic resistance genes affected by soil minerals. Eur. J. Soil Sci. 2020, 72, 1143–1153.
- Liu, X.M.; Wang, D.; Wang, L.; Tang, J.C. Dissolved biochar eliminates the effect of Cu(II) on the transfer of antibiotic resistance genes between bacteria. J. Hazard. Mater. 2022, 424, 127251.
- Zheng, H.; Wang, R.R.; Zhang, Q.; Zhao, J.; Li, F.; Luo, X.; Xing, B.S. Pyroligneous acid mitigated dissemination of antibiotic resistance genes in soil. Environ. Int. 2020, 145, 106158.
- Yu, K.Q.; Chen, F.R.; Yue, L.; Luo, Y.; Wang, Z.; Xing, B. CeO2 nanoparticles regulate the propagation of antibiotic resistance genes by altering cellular contact and plasmid transfer. Environ. Sci. Technol. 2020, 54, 10012–10021.
- Liu, X.M.; Wang, D.; Tang, J.C.; Liu, F.; Wang, L. Effect of dissolved biochar on the transfer of antibiotic resistance genes between bacteria. Environ. Pollut. 2021, 288, 117718.
- Zhou, G.W.; Zheng, F.; Fan, X.T.; Li, M.-J.; Sun, Q.-Y.; Zhu, Y.-G.; Yang, X.R. Host age increased conjugal plasmid transfer in gut microbiota of the soil invertebrate Caenorhabditis elegans. J. Hazard. Mater. 2022, 424, 127525.
- Beaber, J.W.; Hochhut, B.; Waldor, M.K. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 2004, 427, 72–74.
- Lu, Y.; Zeng, J.M.; Wu, B.N.; Shunmei, E.; Wang, L.; Cai, R.; Zhang, N.; Li, Y.; Huang, X.; Huang, B.; et al. Quorum sensing N-acyl homoserine lactones-SdiA suppresses Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting tral expression. Front. Cell. Infect. Microbiol. 2017, 7, 7.
- Zheng, J.; Chen, T.; Chen, H. Antibiotic resistome promotion in drinking water during biological activated carbon treatment: Is it influenced by quorum sensing? Sci. Total Environ. 2018, 612, 1–8.
- Lu, W.W.; Wang, M.; Wu, J.Q.; Jiang, Q.Y.; Jin, J.R.; Jin, Q.; Yang, W.W.; Chen, J.; Wang, Y.J.; Xiao, M. Spread of chloramphenicol and tetracycline resistance genes by plasmid mobilization in agricultural soil. Environ. Pollut. 2020, 260, 113998.
- van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottova, D.; Kristufek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164.
- Chen, Q.L.; An, X.L.; Li, H.; Zhu, Y.G.; Su, J.Q.; Cui, L. Do manure-borne or indigenous soil microorganisms influence the spread of antibiotic resistance genes in manured soil? Soil Biol. Biochem. 2017, 114, 229–237.
- Van Acker, H.; Coenye, T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017, 25, 456–466.
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 3–8.
- Reniere, M.L. Reduce, Induce, Thrive: Bacterial Redox Sensing during Pathogenesis. J. Bacteriol. 2018, 200, e00128-18.
- Chen, X.F.; Yin, H.L.; Li, G.Y.; Wang, W.; Wong, P.K.; Zhao, H.; An, T. Antibiotic-resistance gene transfer in antibiotic-resistance bacteria under different light irradiation: Implications from oxidative stress and gene expression. Water Res. 2019, 149, 282–291.
- Han, X.; Lv, P.; Wang, L.G.; Long, F.; Ma, X.-L.; Liu, C.; Feng, Y.-J.; Yang, M.-F.; Xiao, X. Impact of nano-TiO2 on horizontal transfer of resistance genes mediated by filamentous phage transduction. Environ. Sci.-Nano 2020, 7, 1214–1224.
- Sagar, S.; Kumar, R. Role of SOS response in bacterial drug resistance. Int. J. Pharm. Sci. Rev. Res. 2014, 25, 102–105.
- Ding, C.S.; Pan, J.; Jin, M.; Yang, D.; Shen, Z.; Wang, J.; Zhang, B.; Liu, W.; Fu, J.; Guo, X.; et al. Enhanced uptake of antibiotic resistance genes in the presence of nanoalumina. Nanotoxicology 2016, 10, 1051–1060.
- Tan, H.D.; Fu, L.; Seno, M. Optimization of bacterial plasmid transformation using nanomaterials based on the yoshida effect. Int. J. Mol. Sci. 2010, 11, 4962–4973.
- Hernandez, G.; Bollini, A.; Huarte, M.; Bazzoni, G.; Piehl, L.; Chiarotto, M.; de Celis, E.R.; Rasia, M. In vitro effect of aluminium upon erythrocyte membrane properties. Clin. Hemorheol. Microcirc. 2008, 40, 191–205.
- Yamamoto, Y.; Hachiya, A.; Matsumoto, H. Oxidative damage to membranes by a combination of aluminum and iron in suspension-cultured tobacco cells. Plant Cell Physiol. 1997, 38, 1333–1339.
- Ma, W.T.; Peng, D.H.; Walker, S.L.; Cao, B.; Gao, C.-H.; Huang, Q.; Cai, P. Bacillus subtilis biofilm development in the presence of soil clay minerals and iron oxides. Npj Biofilms Microbiomes 2017, 3, 4.
- Ouyang, K.; Walker, S.L.; Yu, X.Y.; Gao, C.-H.; Huang, Q.; Cai, P. Metabolism, survival, and gene expression of Pseudomonas putida to hematite nanoparticles mediated by surface-bound humic acid. Environ. Sci.-Nano 2018, 5, 682–695.
- Wang, Q.; Liu, L.; Hou, Z.L.; Wang, L.; Ma, D.; Yang, G.; Guo, S.; Luo, J.; Qi, L.; Luo, Y. Heavy metal copper accelerates the conjugative transfer of antibiotic resistance genes in freshwater microcosms. Sci. Total Environ. 2020, 717, 137055.
- Li, M.Z.; He, Y.N.; Sun, J.; Li, J.; Bai, J.; Zhang, C. Chronic exposure to an environmentally relevant triclosan concentration induces persistent triclosan resistance but reversible antibiotic tolerance in Escherichia coli. Environ. Sci. Technol. 2019, 53, 3277–3286.
- Chen, I.; Christie, P.J.; Dubnau, D. The ins and outs of DNA transfer in bacteria. Science 2005, 310, 1456–1460.
- Samuels, A.L.; Lanka, E.; Davies, J.E. Conjugative junctions in RP4-mediated mating of Escherichia coli. J. Bacteriol. 2000, 182, 2709–2715.
- Zhong, X.; Krol, J.E.; Top, E.M.; Krone, S.M. Accounting for mating pair formation in plasmid population dynamics. J. Theor. Biol. 2010, 262, 711–719.
- Diethmaier, C.; Chawla, R.; Canzoneri, A.; Kearns, D.B.; Lele, P.P.; Dubnau, D. Viscous drag on the flagellum activates Bacillus subtilis entry into the K-state. Mol. Microbiol. 2017, 106, 367–380.
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020, 28, 668–681.
- Tsuneda, S.; Aikawa, H.; Hayashi, H.; Yuasa, A.; Hirata, A. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS Microbiol. Lett. 2003, 223, 287–292.
- Hu, X.J.; Kang, F.X.; Yang, B.; Zhang, W.; Qin, C.; Gao, Y. Extracellular polymeric substances acting as a permeable barrier hinder the lateral transfer of antibiotic resistance genes. Front. Microbiol. 2019, 10, 736.
- Xayarath, B.; Freitag, N.E. When being alone is enough: Noncanonical functions of canonical bacterial quorum-sensing systems. Future Microbiol. 2016, 11, 1447–1459.
- Raju Maddela, N.; Sheng, B.; Yuan, S.; Zhou, Z.; Villamar-Torres, R.; Meng, F. Roles of quorum sensing in biological wastewater treatment: A critical review. Chemosphere 2019, 221, 616–629.
- Shank, E.A.; Kolter, R. Extracellular signaling and multicellularity in Bacillus subtilis. Curr. Opin. Microbiol. 2011, 14, 741–747.
- Inaoka, T.; Ochi, K. Re1A protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 2002, 184, 3923–3930.
- Lopez, D.; Vlamakis, H.; Kolter, R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 2009, 33, 152–163.
- Seitz, P.; Blokesch, M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental gram-negative bacteria. FEMS Microbiol. Rev. 2013, 37, 336–363.
- Vestergard, M.; Ekelund, F.; Winding, A.; Jacobsen, C.S.; Christensen, S. Starved bacteria retain their size but lose culturability-Lessons from a 5000 years old undisturbed A-horizon. Soil Biol. Biochem. 2011, 43, 1379–1382.
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J.-P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 2014, 12, 181–196.
- Lu, N.; Zilles, J.L.; Nguyen, T.H. Adsorption of extracellular chromosomal DNA and its effects on natural transformation of Azotobacter vinelandii. Appl. Environ. Microbiol. 2010, 76, 4179.
- Pei, R.T.; Gunsch, C.K. Plasmid conjugation in an activated sludge microbial community. Environ. Eng. Sci. 2009, 26, 825–831.
- Dahlberg, C.; Bergstrom, M.; Hermansson, M. In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl. Environ. Microbiol. 1998, 64, 2670–2675.
- Bandyopadhyay, A.; O’Brien, S.; Frank, K.L.; Dunny, G.M.; Hu, W.S. Antagonistic donor density effect conserved in multiple Enterococcal conjugative plasmids. Appl. Environ. Microbiol. 2016, 82, 4537–4545.
