The ancient guanine nucleotide-binding (G) proteins are a group of critical regulatory and signal transduction proteins, widely involved in diverse cellular processes of all kingdoms of life. YchF is a kind of universally conserved novel unconventional G protein that appears to be crucial for growth and stress response in eukaryotes and bacteria. YchF is able to bind and hydrolyze both adenine nucleoside triphosphate (ATP) and guanosine nucleoside triphosphate (GTP), unlike other members of the P-loop GTPases.
- YchF
- growth
- stress response
- P-loop NTPase
1. Structural Characterization of G Domain of G Proteins

2. Structural Characterization of G Domain of YchF
|
Homolog |
Species |
Residues |
Location |
Supportive Reasons/Effects |
Functions |
References |
|---|---|---|---|---|---|---|
|
E. coli YchF |
Escherichia coli |
His114 |
A highly flexible loop of G domain |
Supporting the flexible loop to reach a catalytically active conformation |
Critical for ATPase activity (+) |
[3] |
|
E. coli YchF |
Escherichia coli |
Cys35 |
G2 motif |
Allows YchF dimerization via a disulfide bridge |
Critical for ATPase activity (−) |
[4] |
|
E. coli YchF |
Escherichia coli |
Lys78 (Arg) |
G domain |
YchF-K78A mutant shows similar hydrolysis activities in presence of Na+ or K+, but K78R mutant retained potassium specific stimulation of ATPase activity |
Plays a key role in determining the potassium dependent ATPase activity |
[5] |
|
hOLA1 |
Homo sapiens |
Leu96 |
G domain (next to G3 motif) |
A conserved Gln residue involved in GTP hydrolysis in Ras-like GTPases has been replaced |
Inactivates Ras-like GTPases |
[6] |
|
E. coli YchF |
Escherichia coli |
Ser16 (Ser36 in H. sapiens) |
G1 motif |
Ser16 phosphorylated when H2O2 absence; Dissociation of KatG |
Supports the ATPase activity; Detoxifies H2O2 |
|
|
E. coli YchF |
Escherichia coli |
Leu76 |
G3 motif |
Hallmark for HAS-NTPase |
Slightly affects ATPase activity (+) |
[3] |
|
hOLA1 |
Homo sapiens |
Thr37 |
G domain |
The main chain amide of Thr37 contacts the α-phosphate of AMPPCP |
Supports the ATPase activity |
[6] |
|
hOLA1 |
Homo sapiens |
Ser36/Val33 |
G1 motif |
The main chain amide of Ser36 and Val33 contacts the β-phosphate of AMPPCP |
Supports the ATPase activity |
[6] |
|
hOLA1 |
Homo sapiens |
Asn32 |
G1 motif |
The main chain amide of Asn32 forms a hydrogen bond to the γ-phosphate of AMPPCP |
Supports the ATPase activity |
[6] |
|
hOLA1 |
Homo sapiens |
Asn230 |
G4 motif |
Its mutation to alanine abolished nucleotide binding |
Contribute to nucleotide binding |
[6] |
|
hOLA1 |
Homo sapiens |
Leu231 |
G4 motif |
Specificity for adenine binding is based on the interaction between the adenine N-6 group and Leu231 main chain CO in G4 motif |
Make YchF preference for ATP rather than GTP |
[6] |
|
hOLA1 |
Homo sapiens |
Ser310 |
TGS domain |
The H-bond between Ser310 Oγ and the exocyclic N-6 of an adenine is formed in a position similar to the ppGpp O-6 |
Make YchF preference for ATP rather than GTP |
[6] |
|
hOLA1 |
Homo sapiens |
Phe127 |
Coiled-coil domain |
Mutating this residue to Ala diminishes ATP binding drastically |
Contribute to base recognition |
[6] |
|
AtYchF1 |
Arabidopsis thaliana |
Glu345 |
TGS domain |
Conserved and solvent-exposed |
Most critical for its interaction with the regulator, GAP1 |
[8] |
Amino acids in brackets indicate that there are other known residues presenting at the same position in the orthologs; In the “functions” column, (+) means upregulation, and (−) means down-regulation.
3. Structural Comparison of G Domains among Selected YchF, Small G Protein, and Heterotrimeric G Protein α-Subunit
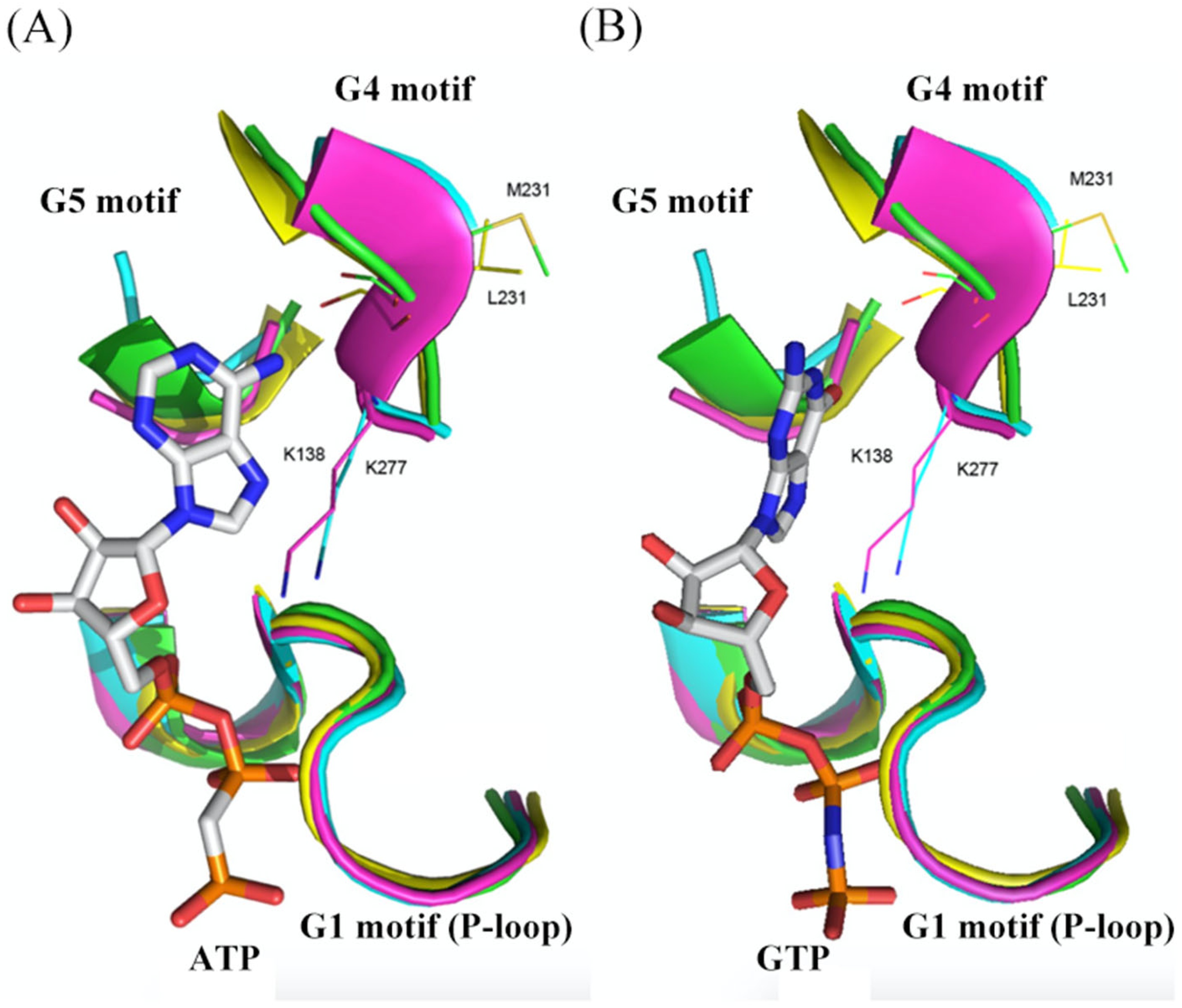
This entry is adapted from the peer-reviewed paper 10.3390/life13041058
References
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118 Pt 5, 843–846.
- Luo, M.; Han, Z.; Huang, G.; Li, R.; Liu, Y.; Lu, J.; Liu, L.; Miao, R. Structural comparison of unconventional G protein YchF with heterotrimeric G protein and small G protein. Plant Signal. Behav. 2022, 17, 2024405.
- Rosler, K.S.; Mercier, E.; Andrews, I.C.; Wieden, H.J. Histidine 114 Is Critical for ATP Hydrolysis by the Universally Conserved ATPase YchF. J. Biol. Chem. 2015, 290, 18650–18661.
- Hannemann, L.; Suppanz, I.; Ba, Q.; MacInnes, K.; Drepper, F.; Warscheid, B.; Koch, H.G. Redox Activation of the Universally Conserved ATPase YchF by Thioredoxin 1. Antioxid. Redox Signal. 2016, 24, 141–156.
- Tomar, S.K.; Kumar, P.; Prakash, B. Deciphering the catalytic machinery in a universally conserved ribosome binding ATPase YchF. Biochem. Biophys. Res. Commun. 2011, 408, 459–464.
- Koller-Eichhorn, R.; Marquardt, T.; Gail, R.; Wittinghofer, A.; Kostrewa, D.; Kutay, U.; Kambach, C. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J. Biol. Chem. 2007, 282, 19928–19937.
- Wenk, M.; Ba, Q.; Erichsen, V.; MacInnes, K.; Wiese, H.; Warscheid, B.; Koch, H.G. A universally conserved ATPase regulates the oxidative stress response in Escherichia coli. J. Biol. Chem. 2012, 287, 43585–43598.
- Cheung, M.Y.; Ngo, J.C.; Chen, Z.; Jia, Q.; Li, T.; Gou, Y.; Wang, Y.; Lam, H.M. A structure model explaining the binding between a ubiquitous unconventional G-protein (OsYchF1) and a plant-specific C2-domain protein (OsGAP1) from rice. Biochem. J. 2020, 477, 3935–3949.
- Sun, H.; Luo, X.; Montalbano, J.; Jin, W.; Shi, J.; Sheikh, M.S.; Huang, Y. DOC45, a novel DNA damage-regulated nucleocytoplasmic ATPase that is overexpressed in multiple human malignancies. Mol. Cancer Res. 2010, 8, 57–66.
