The protein acylation of either the α-amino groups of amino-terminal residues or of internal lysine or cysteine residues is one of the major posttranslational protein modifications that occur in the cell with repercussions at the protein as well as at the metabolome level. The lysine acylation status is determined by the opposing activities of lysine acetyltransferases (KATs) and lysine deacetylases (KDACs), which add and remove acyl groups from proteins, respectively. A special group of KDACs, named sirtuins, that require NAD+ as a substrate has received particular attention in recent years. They play critical roles in metabolism, and their abnormal activity has been implicated in several diseases. Conversely, the modulation of their activity has been associated with protection from age-related cardiovascular and metabolic diseases and with increased longevity. The benefits of either activating or inhibiting these enzymes have turned sirtuins into attractive therapeutic targets, and considerable effort has been directed toward developing specific sirtuin modulators.
- posttranslational modifications
- protein acylation
- lysine deacetylases
- sirtuins
1. Introduction
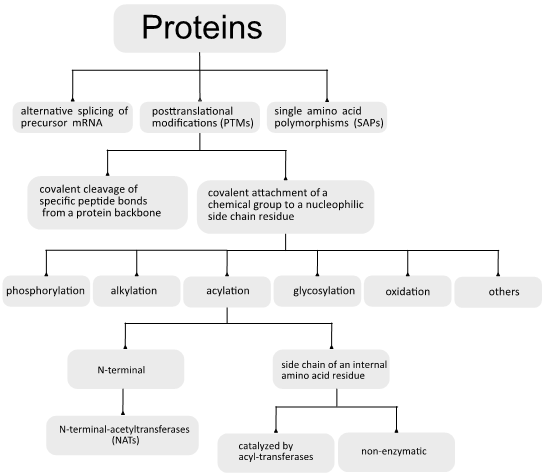
Proteins are the structural and functional base of all living organisms. To date, the number of proteins that comprise the human proteome is still elusive. The analysis of the human genome shows the existence of approximately 25,000 protein-coding genes [1]. This would suggest the existence of the same number of proteins. Interestingly, to date, more than 90,000 different human proteins have been identified. This discrepancy has been attributed to three district mechanisms: the alternative splicing of precursor mRNAs, single amino acid polymorphisms (SAPs), and posttranslational modifications (PTMs) [2–4]. Together, these modifications raise the complexity of the proteomes by two to three orders of magnitude and help to explain the discrepancy between the complexity of vertebrate organisms and the sizes of their encoded genomes [2,5].
There are two general categories of protein PTM: the covalent cleavage of specific peptide bonds from a protein backbone, which can occur by autocatalytic cleavage or can be catalyzed by a group of enzymes named proteases, and the covalent attachment of a chemical group, which is usually an electrophilic fragment of a co-substrate, to a nucleophilic side-chain residue from the protein. The last reaction can be reversible or irreversible and can be enzymatically or non-enzymatically catalyzed.
It has been estimated that about 5% of the genomes of higher eukaryotes are dedicated to enzymes involved in the posttranslational modification of proteins [2]. Currently, there is a record of more than 300 different PTMs [2,6], and the most common include phosphorylation, ubiquitination, alkylation, glycosylation, oxidation, and acylation [5,7–11].
The variability that these PTMs bring to proteins is further amplified by the existence of complex regulatory networks involving both positive and negative crosstalk between the different PTMs. This complex PTM crosstalk is the basis of a protein modification code and has significant importance in the regulation of cellular functions [5,12–14].
The most extensively studied PTMs are phosphorylation and acylation [9,11]. Among the array of different possible acylation reactions that may occur, acetylation is the most common (for example, more than 20% of mitochondrial proteins are acetylated [15]) [11,13,16–18].
The function of protein acylation reactions depends on the acyl group that is attached. It was proposed that the small acyl groups (formyl and acetyl) may function as recognition elements for protein–protein interactions, while long chains of fatty acids can target proteins to membranes and affect signal transduction [19].
The aim of this review is to provide a global overview of the mechanisms underlying both the acylation (Figure 1) and deacylation (Figure 2) of proteins, focusing on a special class of NAD+-dependent protein deacylases named sirtuins.
Figure 1. General overview of the cellular events related with the protein acylation process.
Figure 2. Classification of different lysine deacylases (KDACs) based on their phylogenetic conservation and/or sequence similarities.
2. Protein Acylation
Protein acylation consists of the covalent attachment of acyl groups (acetyl, propionyl, butyryl, malonyl, succinyl, crotonyl, 2-hydroxyisobutyryl, glutaryl, benzoyl, myristate, palmitate, farnesyl, geranylgeranyl, etc.) to amino acid residues bearing nucleophilic side chains, such as lysine (-NH2), cysteine (-SH), serine and threonine (-OH), with a specific location in the target protein [19].
The acyl group is generally attached to the side chain of an internal amino acid residue, but acylation can also occur in the protein N-terminus.
2.1. N-Terminal Acylation
The known examples of N-terminal acylation are the N-myristoylation of glycine, the N-palmitoylation of cysteine, and N-terminal acetylation [19]. Among those, the most common and best characterized is N-terminal acetylation [20].
N-Terminal Acetylation
In eukaryotes, the acetylation of an α-amino group of the N-terminus is an irreversible covalent modification catalyzed by N-terminal-acetyltransferases (NATs) that occurs co-translationally in more than 80% of all proteins [21,22]. N-terminal acetylation neutralizes the positive charge of the free amino group and blocks the N-terminus from further modifications. This modification influences several protein characteristics such as folding, lifetime and degradation, subcellular localization, interactions and complex formation [23,24]. Eukaryotes possess different NATs with different substrate specificities (NatA, NatB, NatC, NatD, NatE and NatF) [25]. Depending on the NAT isoform, the acetyl group may be transferred to the non-cleaved initial methionine residue of the nascent polypeptide chain, or the reaction may occur after the excision of the methionine residue by the ribosomally bound methionine aminopeptidases, and the acetyl group is added to the residue that is positioned immediately after the excised methionine.
2.2. Acylation of Internal Lysine Residues
The acylation of internal lysine residues is a reversible and highly regulated PTM that targets many proteins from different cellular compartments, such as the mitochondria, cytosol, nucleus and lumen of the endoplasmic reticulum (ER) [26–28]. The target proteins are involved in distinct cellular processes, such as the cell cycle, nuclear transport, chromatin remodeling, mRNA splicing, actin nucleation, signal transduction [26], protein homeostasis and autophagy [27,28].
Although protein acylation can be an activating or inhibitory modification, recent metabolic studies have suggested that, in mitochondria, acylation has a consistently inhibitory role [29,30]
Protein acylation may occur by two distinct mechanisms: it may be catalyzed by acyl-specific transferases that transfer acyl groups from acyl-coenzyme A (acyl-CoA) to the amino acid residues [24,31–33], or it may occur non-enzymatically due to the intrinsic reactivity of acyl-CoA thioesters [29,33–35].
The first evidence of acylation as a PTM came in 1964 when lysine acetylation was discovered as a PTM of histones [36]. The evidence for the addition of other acyl groups as a PTM was only recently discovered, and therefore, the function and mechanisms behind them are still elusive [7].
Acetylation of Internal Lysine Residues
Similar to terminal acetylation, the addition of an acetyl group to an internal lysine residue increases the size of its side chain and neutralizes the positive charge of its amino group. This modification will raise the protein hydrophobicity and consequently induce significant conformational changes that will ultimately affect several protein properties, including transcriptional activity, DNA–protein interactions, subcellular localization, enzymatic activity, folding, peptide–receptor recognition and protein stability [15].
Enzymatic Nε-lysine acetylation has been deeply studied and requires an acetyl group donor (acetyl-CoA), an acetyl group acceptor (the ε-amino group of an internal lysine residue from a polypeptide chain) and an acetyl-CoA:lysine acetyltransferase (lysine acetyltransferase (KAT)) to catalyze the acetyl exchange. The mechanism behind the enzymatic addition of other acyl groups to Nε-lysine requires further studies [29].
Non-enzymatic protein acetylation was identified decades ago [37], and its occurrence was found to be especially favorable in the mitochondria [38–42].
The particular biochemical properties of the mitochondria, namely, its pH and metabolite concentrations, are proposed to favor the occurrence of other non-enzymatic acylation reactions, namely, succinylation, malonylation and glutarylation [29].
2.3. S-Palmitoylation of Cysteine Residues
The reversible attachment of palmitic acid to an internal cysteine residue (S-palmitoylation) is the most common type of protein fatty acylation in eukaryotic cells [43–45]. The attachment of the palmitic acid to the protein is catalyzed by palmitoyl transferases [46,47], and its removal is mediated by palmitoyl thioesterases.
The palmitoyl transferases belong to the zinc-finger DHHC domain-containing protein family that is characterized by the presence of a conserved aspartate–histidine–histidine–cysteine (DHHC) cysteine-rich domain [46,47]. They are integral membrane proteins, with four to six transmembrane domains and the N and C termini present in the cytosol [48], a characteristic that has made difficult their purification and consequently their identification.
In spite of the recent advances in the field, there are still many questions regarding the functioning of DHHC enzymes. To date, their selectivity, their forms of substate selection and the identification of palmitoylation sites are still elusive [44].
2.4. The Influence of Protein Acylation in Human Health and Disease
The attachment of acyl groups to positively charged lysine residues neutralizes the positive charge and consequently affects the protein’s physicochemical properties, ultimately affecting its stability, function, catalytic activity, protein–protein and protein–DNA interactions, degradation and subcellular location [43,44,49].
Many of these post-translationally acylated proteins have relevant roles in vital physiological processes including gene transcription, cell division, cytoskeleton organization, DNA damage repair, DNA replication, signal transduction, protein folding, autophagy, apoptosis, lipid storage and breakdown, mitochondrial fission and fusion, protein synthesis, ion transport, redox and metabolism regulation. [49] Additionally, protein acylation has also been related to the protein aggregation process, which is implicated in several neurological pathologies such as amyotrophic lateral sclerosis [50] and Alzheimer’s disease [51].
Therefore, the correct functioning and regulation of the enzymes involved in protein acylation (and deacylation) are essential for human health, and their misregulation has been associated with several pathologies.
There is evidence of germline mutations in several KATs (for example, KAT6A, KAT6B, CREB- binding protein (CBP) and EP300) that result in disorders, for example, Say–Barber–Biesecker–Young–Simpson (or Ohdo) syndrome [52], Genitopatellar syndrome [53,54] or Rubinstein–Taybi syndrome. These and other disorders caused by KAT mutations are associated with intellectual impairment, developmental delays and physical abnormalities such as facial dysmorphisms [52–56].
Protein acetylation has been associated with several cancers [57], inflammation, immunity (Falkenberg, 2014, pp. 673–691) and metabolic diseases such as diabetes [58].
In spite of its evident therapeutic potential, the inhibition of KATs has not been widely explored. A recent study described the development of A-485, a potent and selective catalytic inhibitor of p300 and CREB-binding protein (CBP) that competes with the substrate acetyl-CoA. The compound selectively inhibited cell proliferation in lineage-specific tumor types, including several hematological malignancies and androgen receptor-positive prostate cancer [59].
Another study reported the development of two highly potent and selective inhibitors of KAT6A and KAT6B named WM-8014 and WM-1119, respectively, that are reversible competitors of acetyl-CoA. The inhibition of KAT6A and KAT6B inhibited the growth of lymphoma in mice [60].
Detailed knowledge about the catalytic activity of the different KATs would be important for identifying potent and selective inhibitors for other KATs that would enable obtaining deep knowledge about their biological functions and exploring their therapeutic potential.
Reversible S-palmitoylation is a dynamic process that has several important functions in subcellular protein trafficking, in protein stability (by the prevention of ubiquitination and subsequent degradation) and in the modulation of protein interactions (adhesion and signaling), but its most studied function is its capability to increase the affinity of soluble proteins for lipophilic membranes [61].
The alteration of DHHC expression and the consequent palmitoylation impairment have been associated with several cancers (for example, leukemia, colorectal, hepatocellular and non-small-cell lung cancers) [44,62,63] and with several other disease states resulting from organ-specific processes. One of the most studied cases is the relation between palmitoylation and neuronal functions. There are many reports relating defects in palmitoylation regulation or in the enzymes responsible for palmitoylation and depalmitoylation processes with several neurological disorders such as Alzheimer’s, Parkinson’s or Huntington’s disease, schizophrenia and intellectual disability [44,64].
Protein Acylation and “Carbon Stress”
It has been recently suggested that an increase in the concentration of reactive carbon metabolites, resulting from physiological or pathological situations, can culminate in the abnormal occurrence of non-enzymatic protein acylation reactions [29]. This situation can have a detrimental effect on protein function and ultimately disrupt cellular homeostasis. In these scenarios, the non-enzymatic protein acylations are considered a form of “carbon stress” [29].
Recently, it has been found that under carbon stress situations that lead to an increase in protein acylation, a particular group of deacylases, named sirtuins (Sirts), are called to intervene, and, in some conditions, their expression can be upregulated. The sirtuins are part of a protein quality control response that is vital for reducing protein acylation, ensuring protein quality control and consequently reducing carbon stress. An increase in carbon stress and/or the impairment of the carbon stress response will reduce protein quality control and ultimately reduce protein function, leading to several age-related diseases such as neurodegeneration, diabetes, cardiovascular disease or cancer [18,65–70].
3. Protein Deacylation
Efficient and correct deacetylation activity is of major importance for the correct functioning of several vital biological processes, including DNA recombination [71] and repair [72], transcriptional silencing [73], axonal protection [74], fat mobilization [75], apoptosis [76,77] and aging [78,79].
The hydrolysis of acyl groups from acyl-lysine residues is catalyzed by a group of enzymes named lysine deacetylases (KDACs).
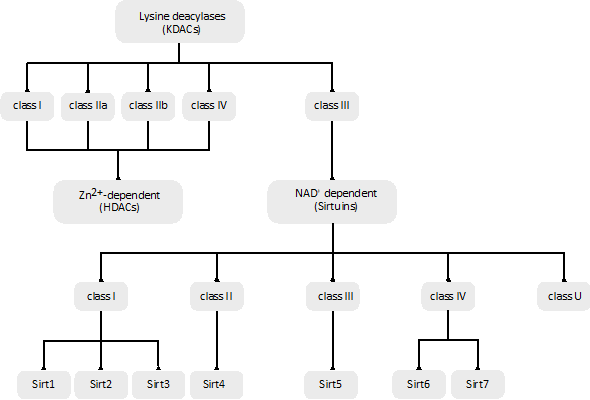
The human genome encodes a total of 18 KDACs that can be grouped into two categories or superfamilies: the Zn2+-dependent KDACs or histone deacetylases (HDACs) and the NAD+-dependent sirtuin deacetylases (Figure 2).
It should be noted that although grouped in the same family, some KDACs have different acyl selectivities [80–83], and others are thought to have poor or no deacetylase activity [84].
3.1. Zn2+-Dependent KDACs
The Zn2+-dependent KDACs (or HDACs) are usually called the classic KDACs and account for 11 of the 18 KDACs. They share a highly conserved deacetylase domain and, based on their phylogenetic conservation and in-sequence similarities, can be divided into four classes, named classes I, IIa, IIb and IV (Figure 2). They differ in their enzymatic function, structure, expression patterns and subcellular localization. Classes I and IV are nuclear, class IIb is cytoplasmic, and class IIa is primarily nuclear but upon the activation of signaling is exported to the cytoplasm [84]. Some in vitro experiments suggest that vertebrate class IIa KDACs show poor catalytic activity, which may be related to the replacement of a conserved tyrosine by a histidine in the catalytic pocket [85].
These enzymes exist in multiprotein complexes that determine their substrate specificity, and their catalysis usually generates deacetylated lysine and acetate [49,84,86].
3.2. NAD+-Dependent Sirtuin Deacetylases
The remaining seven KDACs are named sirtuins and unlike the classic KDACs, they require the cofactor NAD+ as a co-substrate [87].
The name sirtuin came from the family’s founding member, the silent information regulator 2 (Sir2) from Saccharomyces cerevisiae [88].
They comprise the class III family of KDACs and have seven members (Sirt1–Sirt7). Sirtuins are evolutionarily conserved in all domains of life, and, based on their sequence similarity, they are classified into five classes (I–IV and U) (Figure 2). The seven mammalian sirtuin genes are included in classes I to IV: Sirt1, Sirt2 and Sirt3 are in class I, Sirt4 in class II, Sirt5 in class III, and Sirt6 and Sirt7 in class IV (Figure 2) [89]. Sirt1 has the highest sequence homology to Sir2 in yeast, and it is the most studied sirtuin. The different sirtuins are distinguished by their subcellular localization, acyl lysine substrate specificity, enzymatic activity, and biochemical and metabolic functions. Their impairment is related to different metabolic and health issues [90].
Although deacylation is the main activity catalyzed by the sirtuin enzymes, there is evidence that some sirtuins (Sirt1, 4 and 6) can also ADP-ribosylate protein substrates [91].
This entry is adapted from the peer-reviewed paper 10.3390/ijms21228609