Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Triple-negative breast cancer (TNBC) is a more aggressive type of breast cancer due to its heterogeneity and complex molecular mechanisms. TNBC has a high risk for metastasis, and it is difficult to manage clinical conditions of the patients. Long non-coding RNAs (lncRNAs) have emerged as a novel target to treat the multistep process of TNBC. LncRNAs regulate epigenetic expression levels, cell proliferation and apoptosis, and tumour invasiveness and metastasis. Thus, lncRNA-based early diagnosis and treatment options could be helpful, especially for patients with severe TNBC.
- triple-negative breast cancer
- lncRNA
- diagnosis
- targeted drug development and resistance
1. LncRNAs
lncRNAs are actively involved in gene expression, epigenetic deregulation, chromatin remodelling, DNA methylation, translation of oncogenic gene targets, and biogenesis (Figure 1). They are transcribed by RNA polymerase II, after which most transcripts are spliced, and are mainly found in the nucleus and chromatin, being expressed in cells and tissues in a specific manner [6,9,17]. Transcriptional regulation and various molecular processes in the cytoplasm are controlled by lncRNAs; various circulating lncRNAs are transmitted via exosomes and bind to various transcription factors, chromatin-regulated complexes, RNA-binding proteins, nascent RNA transcripts, and chromatin [17]. The normal expression of lncRNAs and the effect of their expression changes on tumour behaviour depends on the canonical function of the mRNA target genes (Figure 2). lncRNAs can bind to the active site of proteins and regulate molecular processes at the post-transcriptional level. They are involved in functional biological processes at the cellular or physiological levels. RNA-induced silencing complexes (RISCs) are formed with the help of lysine-specific demethylase 5B (KDM5B, also known as histone demethylase JARID1B), trimethylation of lysine 4 on the histone H3 protein subunit (H3K4me3), monomethylation of lysine 4 on the histone H3 protein subunit (H3K4me1), hsa-miR-448 (also known as miRNA448), breast cancer 1/2 (BRCA1/2), retinoblastoma protein (pRB), caveolin-1 (CAV-1), Homeobox protein Hox-A5 (HOXA5), Stratifin (SFN), methyl groups (CH3), and Ras homolog gene family, member A (RhoA) (Figure 1 and Figure 3) [18]. In 2019, it was found that the lncRNA MIR100HG regulates proliferation in TNBC and the expression of the p27 gene after formation of an RNA–DNA triplex at the promoter [19]. Moreover, MIR100HG silencing leads to reduced transcription and translation of p27 [19,20]. Three triplex-forming oligonucleotides (TFOs) have been observed on the lncRNA of p27, which binds to the triplex-targeting ability (TTA) site at the 5’UTR; this event has been observed in TNBC cell lysates [21]. The binding of TFO1 and TTA is a unique mechanism by which MIR100HG regulates the transcription factors at the promoter region of p27 [21,22]. Plasmacytoma variant translocation 1 (PVT1) is another type of lncRNA that is transcribed by a gene situated at the 8q24 chromosomal region and plays and important role in TNBC development. It contains 12 exons that when spliced generate lncRNAs [23]. PVT1 binds to Krüppel-like factor 5 (KLF5) and generates a BAP1 deubiquitinase that induces TNBC via beta-catenin upregulation. Furthermore, the PVT1 promoter also acts as a regulator of the expression of the MYC proto-oncogene and BHLH transcription factor (c-MYC) [24]. These findings show that lncRNAs also mediate regulation at the transcriptional level.
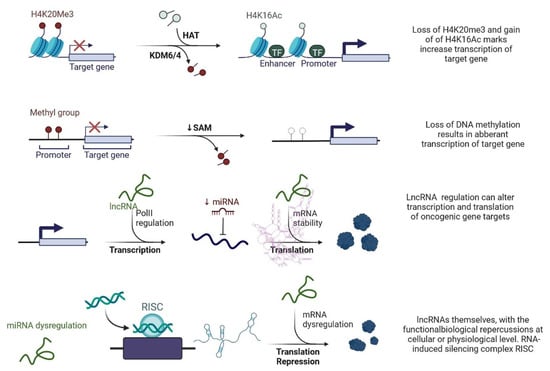
Figure 1. Epigenetic deregulation in cancer including chromatin remodelling, DNA methylation, and non-coding RNA regulation that alters transcription and translation of oncogenic gene targets.
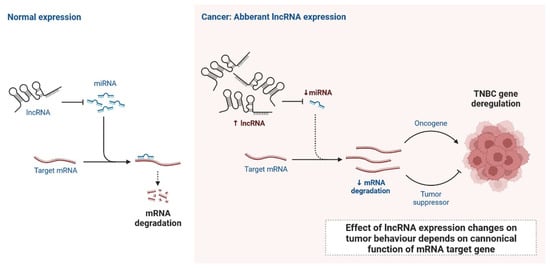
Figure 2. Normal expression of lncRNA and effect of lncRNA expression changes on tumour behaviour depends on canonical function of mRNA target gene.
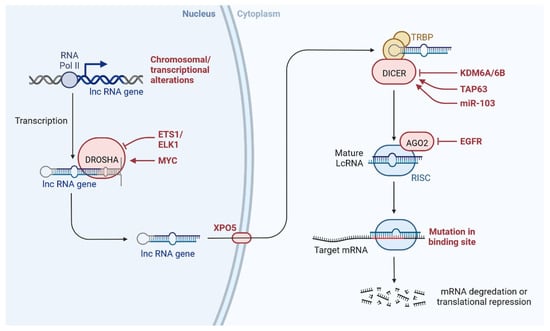
Figure 3. lncRNAs are involved with the functional repercussions at the cellular and physiological level. RNA-induced silencing complex (RISC): KDM5B (lysine-specific demethylase 5B also known as histone demethylase JARID1B), H3K4me3 (trimethylation of lysine 4 on the histone H3 protein subunit), H3K4me1 (monomethylation of lysine 4 on the histone H3 protein subunit), hsa-miR-448 (also known miRNA448), BRCA1/2 (breast cancer 1/2), pRB (retinoblastoma protein), CAV 1 (caveolin 1), HOXA5 (Homeobox protein Hox-A5), SFN (Stratifin), CH3 (methyl group), and RhoA (Ras homolog gene family, member A).
2. Clinical Updates on lncRNAs in TNBC
Recently, lncRNA expression in patients with TNBC was investigated; 1034 lncRNAs were identified using NGS technologies and microarrays, out of which, 537 lncRNAs regulate 451 protein-coding genes [14]. These genes are also detected in TNBC cells and are involved in cell signalling pathways such as the MAPK and PI3K-Akt pathways, which may lead to heterogeneity [14,24]. lncRNAs also act as miRNAs, binding to miRNA-targeted mRNAs and dysregulated miRNAs [25]. This crosstalk forms a complex post-transcriptional regulatory network including mRNAs and lncRNAs that is called the competing endogenous RNA (ceRNA) network [26]. ceRNA-mediated regulatory mechanisms constitute an important pathway in lncRNA-modulated post-transcriptional regulation in TNBC [27]. A microarray-based ceRNA network analysis revealed that 4852 lncRNAs are related to the diagnosis and treatment outcome of TNBC [28]. Another study using the TCGA database found that 150 lncRNAs are expressed at the tissue level and 823 in serum and these lncRNAs could act as prognostic factors in TNBC [29]. Furthermore, the study found that the lncRNA OSTN-AS1 is a novel immune-related prognostic marker [29]. An integrated ceRNA network involving three miRNAs (CHRDL1, FCGR1A, and RSAD2) and two lncRNAs (HIF1A-AS2 and AK124454) was developed using microarray analysis [30]. These findings demonstrate that lncRNAs play major roles in the regulation of cell signalling, genetic heterogeneity, TNBC development, and pathological features (Figure 4) shown in Table 1.
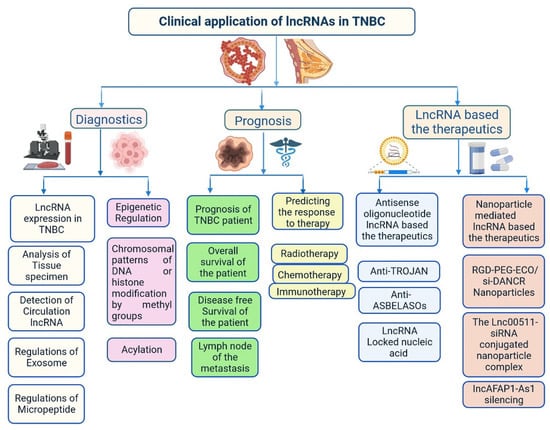
Figure 4. Clinical importance of lncRNA in triple-negative breast cancer.
Table 1. Important lncRNAs associated with triple-negative breast cancer.
| S. N. | lncRNAs | Regulation of Expression | Clinical Importance | Potential Targets | Reference |
|---|---|---|---|---|---|
| 1 | HOTAIR | Upregulation | Increase cell invasion and migration | LEF1/TCF4 | [31] |
| 2 | DRHC | Downregulation | Reduce cell proliferation | HOTAIR | [32] |
| 3 | LINC01133 | Upregulation | Promote phenotypic features like cell stem cells (CSCs) | KLF4 | [33] |
| 4 | LINC01096 | Upregulation | Encourage cell invasion | miR-3130-3p | [34] |
| 5 | HEIH | Upregulation | Increase cell proliferation and prevent cell death | miR-4458/SOCS1 | [35] |
| 6 | ARNILA | Downregulation | Invasion and metastasis | miR-204/SOX4 | [36] |
| 7 | LINC02095 | Upregulation | Promote cell proliferation | SOX9 | [37] |
| 8 | WT1-AS | Downregulation | Inhibit cell migration and invasion | TGF-β1 | [38] |
| 9 | GAS5 | Downregulation | Promote cell apoptosis | miR-378a-5p/SUFU | [39] |
| 10 | CCAT1 | Upregulation | Encourage cell division | miR-218/ZFX | [40] |
| 11 | ASRPS | Downregulation | Inhibit angiogenesis and cell proliferation | STAT3 | [41] |
| 12 | AND2-AS1 | Downregulation | Inhibit angiogenesis inhibit cell division | RUNX2 | [42] |
| 13 | POU3F3 | Upregulation | Promote cell proliferation and inhibit cell apoptosis | Caspase-9 | [43] |
| 14 | NEF | Downregulation | Inhibit cell migration and invasion | miR-155 | [44] |
| 15 | ZEB2-AS1 | Upregulation | Promote cell proliferation, metastasis, and EMT | ZEB2 | [45] |
| 16 | LINC0009 | Upregulation | Increase cell proliferation and invasion | miR-383-5p/RBM3 | [46] |
| 17 | ANRIL | Upregulation | Increase cell proliferation and apoptosis | miR-448/KDM5B | [47] |
| 18 | SNHG12 | Upregulation | Induce cell proliferation, migration, and apoptosis | MMP13 | [48] |
| 19 | LUCAT1 | Upregulation | Encourage cell division, movement, and invasion | miR-5702 | [49] |
| 20 | PCAT6 | Upregulation | Radiotherapy resistance | miR-185-5p/TPD52 | [50] |
| 22 | HULC | Upregulation | Promote metastasis | MMP-2, MMP-9 | [51] |
| 23 | PAPAS | Upregulation | Induce cell migration and invasion | miR-34a | [52] |
| 24 | HCP5 | Upregulation | Increase cell proliferation; reduce cell apoptosis | miR-219a-5p/BIRC3 | [53] |
| 25 | NRAD1 | Upregulation | Stimulate cell proliferation and CSC-like phenotypic traits | miR-219a-5p/BIRC3 | [54] |
| 26 | SNAR | Upregulation | Stimulate cell division | [55] | |
| 27 | AWPPH | Upregulation | Activate cell proliferation | miR-21; FZD7 | [56] |
| 28 | sONE | Downregulation | Prevent cell proliferation | TP53/c-Myc | [57] |
| 29 | DANCR | Upregulation | Promote cell proliferation and invasion | miR-216a-5p | [58] |
| 30 | LINK-A | Upregulation | Increase resistance to immunotherapy, AKT inhibitors, and glycolysis reprogramming | PI3K/GPCR | [59] |
| 31 | MIR503HG | Downregulation | Reduce cell migration and invasion | miR-103/OLFM4 | [60] |
| 32 | NEAT1 | Upregulation | Increase cell apoptosis | [61] | |
| 33 | PTCSC3 | Downregulation | Prevent cell proliferation | H19 | [62] |
| 34 | NRON | Downregulation | Inhibit cell proliferation | snaR | [63] |
| 35 | TROJAN | Upregulation | Promote cell proliferation and invasion | ZMYND8 | [64] |
| 36 | NAMPT-AS | Upregulation | Increase cell metastasis | miR-548b-3p/NAMPT | [14] |
| 37 | MANCR | Upregulation | Promote cell proliferation; inhibit DNA damage | [65] | |
| 38 | RMST | Downregulation | Prevent cell proliferation | [66] | |
| 39 | SK AI1BC | Upregulation | Increase cell migration and invasion | K AI1 | [67] |
| 40 | ROR | Upregulation | Promote cell invasion and metastasis | miR-145/ARF6 | [68] |
| 41 | AIRN | Downregulation | Inhibit cell migration and invasion | Wnt/β-catenin/mTOR/PI3K | [69] |
| 42 | LINC-ZNF469-3 | Upregulation | Promote cell invasion | miR-574-5p/ZEB1 | [70] |
| 43 | PDCD4-AS1 | Downregulation | Inhibit cell proliferation and migration | PDCD4 | [71] |
| 44 | HOST2 | Downregulation | Inhibit cell proliferation | et-7 b/CDK6 | [72] |
| 45 | BORG | Upregulation | Promote doxorubicin resistance | RPA1 | [73] |
| 46 | PVT1 | Upregulation | Promote cell proliferation and migration, and EMT | p21, KLF5/β-catenin | [24] |
| 47 | H19 | Upregulation | Promote paclitaxel resistance and CSC-like phenotypic traits | Akt | [62] |
| 48 | TP73-AS1 | Downregulation | Promote cell vasculogenic mimicry | miR-490-3p/TWIST1 | [74] |
| 49 | TUG1 | Downregulation | Enhance cisplatin sensitivity | miR-197/NLK | [75] |
| 50 | MIR100HG | Upregulation | Promote cell proliferation | p27 | [76] |
| 51 | LINC01638 | Upregulation | Promote cell proliferation | c-Myc | [77] |
2.1. Importance of lncRNAs in Tumour Invasiveness and Metastasis
Tumour invasion and metastasis explain the severity and mortality rate in patients with TNBC (Figure 6) [78,79]. GAS5 overexpression induces the expression of miR-196a-5p, which activates the FOXO1/PI3K/Akt signalling pathway [80]. TROJAN is a drug that reduces the metastasis burden. Degradation of TROJAN is regulated by ZMYND8, and the ubiquitin–proteasome pathway is involved in this process [81]. CCAT1 activates the migration of TNBC cells via miR-218/ZFX signalling [40]. Various ncRNAs are involved in cell migration and invasion via specific regulatory pathways, including MIR503HG through the miR-103/OLFM4 axis [60], CCAT1 through the dysregulation of the miR-218/ZFX axis [40], AFAP1-AS1 through the activation of Wnt/β-catenin signalling [82], miR-34a through the activation of EMT-associated signalling pathways [83], PAPAS through miR-34a.83 downregulation [52], sONE through sONE/NOS3/NO signalling activation [53], LINC-ZNF469-3 by activating the miR-574-5p/ZEB1 axis [71,78], ZEB2 through the activation of PI3K/Akt/GSK3β/ZEB2 signalling [45], PVT1 by regulating p21 and KLF5/β-catenin signalling [24], ARNILA by mimicking ceRNA for miR-204, AIRN by downregulating Wnt/β-catenin/mTOR/PI3K signalling [36], RMST by downregulating Wnt/β-catenin/mTOR/PI3K signalling [67], and MALAT1 by upregulating miR-129-5p and miR-1/Slug expression [84]. Furthermore, miR-448 and some other lncRNAs play very important roles in invasion and metastasis, including SKAI1BC, HULC, HOTAIR, SNHG12, SNAR, WT1-AS, LINC01096, DANCR, NEF, HIF1A-AS2, LncKLHDC7B, and ROR [30,31,32,38,48,55,58,59,60,61,62,63,64,65,66,67,68,69,85,86].
2.2. Importance of lncRNAs in Clinical Diagnosis
Several studies have found that lncRNAs are involved in the regulation of various transcription factors, epigenetic changes, chromatin remodelling, DNA methylation patterns, alternative splicing, post-translational modifications, and interaction with small peptides. All these events have great importance in the early diagnosis and treatment of patients with TNBC [14,86]. lncRNA expression levels in the blood and tissues of patients with TNBC at different stages has been investigated [14]. Based on reverse transcription quantitative PCR analysis data, the lncRNAs HIF1A-AS2, UCA1, and ANRIL can be used for TNBC detection, with areas under the curve in the range of 0.827–0.840, and a diagnostic accuracy of 0.962 for ANRIL [87]. ANRIL, SOX2OT, and ANRASSF1 are used to differentiate between healthy and TNBC cells. TINCR expression is used to differentiate various histological subtypes of BC, as it is highly expressed in TNBC cells [88]. UCA1 is associated with TNBC, acting as a specific marker for TNBC diagnosis. EZH2 is highly expressed in TNBC tissues and prevents apoptosis by activating the miR-4458/SOCS1 axis [89]. LINC00299 expression is increased in TNBC. Several lncRNAs bind to mRNAs, protecting them and increasing their stability. The oncogenic transcription factor SOX9 is activated by LINC02095 [90]. DANCR interacts with RXRA and activates PI3K/Akt signalling in TNBC [58]. LINC00152 enhances NEDD4-1-facilitated ubiquitination and dysregulation of PTEN protein in TNBC [91]. Cell cycle arrest at the G1 phase is induced by MIR100HG, with p27 binding to RNA–DNA; p27 is a cyclin-dependent kinase (CDK) inhibitor. Cell cycle arrest at the G0/G1 phase is induced by LINC00339 and RMST in TNBC through the miR-377-3p/HOXC6 signalling pathway [19,20,77,92]. GAS5 is actively involved in the inhibition of TNBC cells through its action on miR-196a-5p and miR-378a-5p/SUFU signalling [93]. Further understanding of the roles of all these lncRNAs in TNBC is needed to improve early diagnosis and clinical management of patients. Various genes are targeted by ncRNAs, including LARP7, CDKN1A, KLF2, TIA1, DDX3X, CDK, and QKI [94,95,96,97,98]. An analysis of the TCGA database showed that 1097 lncRNAs are expressed in BC, with 1510 differentially expressed lncRNAs in TNBC cells, 35 plasma lncRNAs in TNBC, and 672 in non-TNBC cells [14]. Some lncRNAs are directly linked to prognosis in TNBC, including FOXCUT, LINC00299, AP000924.1, AC091043.1, AL354793.1, AC010343.3, and FGF10-AS1 [14]. Plasma-specific lncRNAs are also used for diagnosis of TNBC, such as UCA1, ANRIL, and HIF1A-AS2 [30]. lncRNAs associated with lymph node metastasis, such as LINC000173, LINC00096, ZEB2-AS1, HIF1A-AS2, HULC, LUCAT1, SNHG12, MALAT1, HOTAIR, HIF1A-AS2, LINC00096, ADPGK-AS1, and ZEB2-AS1, have also shown importance in diagnosis and prognosis [11,14,30,49].
2.3. Importance of lncRNAs in Treatment
lncRNAs affect the response to treatments such as chemotherapy, immunotherapy, and radiotherapy [99]. H19 is expressed in patients with TNBC during neoadjuvant chemotherapy and is related to effective clinical outcomes. LINK-A expression is linked to response to pembrolizumab treatment in patients with TNBC because its decreased expression reduces CD8+ T-cell infiltration [59]. These lncRNAs act as biomarkers for treatment response in patients with TNBC. LncAFAP1-AS1 expression has been observed in patients with TNBC who received radiotherapy after surgery, and this lncRNA acts as biomarker for radiotherapy [82]. Moreover, lncRNAs are involved in angiogenesis. LINC01133 expression is induced by mesenchymal stem/stromal cells that adjoin TNBC cells [33]. lncRNAs are actively involved in the regulation of cell proliferation and apoptosis as well as drug resistance in TNBC [16,44,47,61,99]. DRHC and HOTAIR inhibit TNBC growth and development [31]. HOTAIR plays a role in the invasion and migration of TNBC cells and is used as a biomarker for TNBC metastasis in circulation and tissues, indicating poor survival and response [31,32]. DRHC inhibits TNBC cell proliferation by downregulating the expression of HOTAIR, whereas HOTAIR does not affect the expression level of DRHC. H19 expression is reduced in TNBC cells, whereas PTCSC3 expression is not altered by H19 overexpression [61]. HIST2H2BC and SNRPEP4 were identified in 165 frozen tissue samples by transcriptome microarrays; these lncRNAs are involved in taxane chemotherapy in patients with TNBC. Increased miR-377-3p expression delays TNBC progression by regulating the inc00339/miR-377-3p/HOXC6 axis and inhibits TNBC proliferation and apoptosis. Therefore, it is used as therapeutic target. HIF1A-AS2 expression is upregulated in TNBC mammary tissue, which is linked to overall survival. HOTAIR is closely associated with androgen receptor expression and used as a therapeutic strategy to prevent metastasis. The miR-199a/FOXP2 pathway is induced by LINC01133 and triggers the proliferation of TNBC cells. Various lncRNAs act as stem cell markers, such as DANCR, LINC01638, LINC-ZNF469-3, NEAT1, NRAD1, and ASRPS [75,87]. Some lncRNAs promote vasculogenic mimicry, providing growth supplementation for tumour formation in TNBC. TP73-AS1, which is activated by the miR-490-3p/TWIST1 pathway, is one example. LINK-A alters glycolysis by mediating HIF1α phosphorylation at Tyr565 and Ser7 [3,16,44,47]. MANCR inhibits DNA damage and prevents disease progression [66]. AWPPH is involved in the prevention of tumourigenesis upon treatment with carboplatin; AWPPH small interfering RNA (siRNA) silencing leads to increased chemosensitivity in TNBC [10,56]. TUG1 induces the expression of miR-197, reduces the activation of WNT signalling, and enhances TNBC cell sensitivity to cisplatin [75]. These findings demonstrate the importance of lncRNAs in the prevention of tumourigenesis. More studies are required to explore lncRNA treatment options. Early studies showed that HOTAIR recruits the polycomb repressive complex 2 to its target genes through the CoREST/REST H3K4 demethylase complex [75].
This entry is adapted from the peer-reviewed paper 10.3390/cells12040674
This entry is offline, you can click here to edit this entry!
