Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
This communication aims at discussing strategies based on developments from nanotechnology focused on the next generation of sequencing (NGS). In this regard, it should be noted that even in the advanced current situation of many techniques and methods accompanied with developments of technology, there are still existing challenges and needs focused on real samples and low concentrations of genomic materials.
- next generation of sequencing (NGS)
- DNA detection
- genotyping
- Enhanced Nanospectroscopy
- Enhanced signaling
- Nanotechnology
1. Introduction
Over the last years, research into nanotechnology has shown to be of major importance in basic and applied science [1][2] such as in precision nanomedicine [3], nano-oncology [4][5], and smart responsive nano-optical platforms [6][7] for targeted detection of biomolecules [8] at different concentration levels. These optical active nanoplatforms could also be incorporated into substrates for devices and miniaturized instrumentation [9]. Functional nanoarchitectures could be designed with particular properties for molecular sensing. Modified metallic nanoparticles such as gold, silver, and materials such as core templates with molecular spacers with controlled dimensions could produce nano-optical platforms for chemical surface modifications (Figure 1) [10].
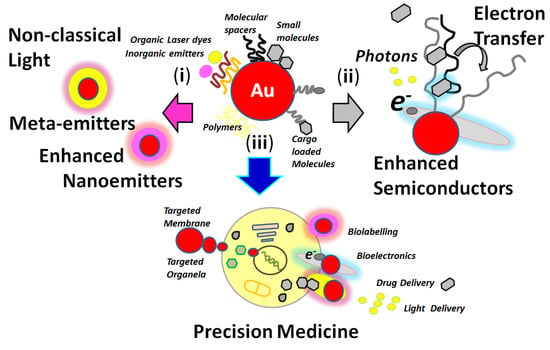
Figure 1. Representation of gold nanoplatforms chemical modifications and targeted studies and applications focused on: (i) nanophotonics; (ii) nanoelectronics; and (iii) nanomedicine. Reprinted with permission from ref. [10].
This is the case of ribonucleic acid (RNA), deoxyribonucleic acid (DNA), and related genomic materials that could interact within the near field and thus develop signal modifications recorded by nanoscopy. From these interactions, varied states of nano-aggregations could lead to new physical phenomena not yet studied as well as those already studied being potentially applied to nanotechnology and the life sciences.
In all cases mentioned, signal detection, transduction, and translation through space and time are required. In addition, the high sensitivity of optical platforms for single-molecule detections (SMD) [11] and different optical setups coupled to the molecular systems under study is well known [12][13]. Therefore, nanotechnology is a multidisciplinary research field that responds to challenges in one of the most significant research areas, namely genomic sequencing [14] and the next generation of sequencing (NGS) [15] in progress. In this ongoing technology, tuned nanomaterials combined with enzymes could develop new strategies for DNA extraction, separation, detection, and quantification with microbeads. Thus, the DNA library was diluted to single-molecule concentration, denatured, and hybridized to individual beads containing sequences complementary to adapt oligonucleotides. Thus, the beads were compartmentalized into water-in-oil micro-vesicles, where clonal expansion of single DNA molecules bound to the beads occurred during emulsion PCR. After amplification, the emulsion was disrupted, and the beads containing clonally amplified template DNA were enriched.
Miniaturization of technology should be underlined to focus analysis and data recording towards individual events such as nucleotide interactions and DNA hybridization. The development of beads and nanoplatforms forming part of more complex functional machineries is applied [16][17].
It is known that sequencing addresses many challenges such as mismatching [18], low concentration in real samples, [19], signal detection after complementary DNA interaction [20], sample handling, clean-up of samples [21], and manipulation and multi-step procedures [22], all varying according to the methodology used. As could be observed, many variables should be controlled to design a new targeted methodology.
2. Advances and Perspectives from Nanotechnology towards NGS
The discussion of perspectives on NGS from the nanomaterial viewpoint is closely related to current nanotechnology developments. From fundamental research with new concepts and designs, proofs of concepts can be discussed and evaluated as derived from real technology. In this perspective, and knowing that NGS is focused on the determination from single nucleotides to the accurate order of variable nucleotide composition within longer DNA or RNA chains [23], researchers should highlighted its potential impact and capability to provide insights within other research fields.
Therefore, there are still challenges to address in relation to the main variables described as well as new ones related to further capabilities using new detection strategies and systems. Researchers show some representative high-impact nanotechnology developments in progress, encouraging innovation in DNA-detection and -genotyping technology.
As known, sequencing requires facing many challenges ranging from low concentrations in real samples and isolation to amplifying genomic material. The amplification procedure increases the quantity of the genomic material by copying targeted sequences. The generation of DNA-strand libraries allows continuing the design of detection strategies. Then, even if many genomic libraries are available, variable epigenetic detection could add extra difficulties. Thus, real samples by intrinsic and natural expression could affect determinations as well, and it should be contemplated in the design of the methodology. In this context, DNA mismatching should be solved. Probably, the most important variable to transduce and generate specific detection signals from single-nucleotide interactions is based on non-covalent forces. Considering natural and non-synthetic nucleotides, DNA detections from short aptamers to longer genomic chains are generally based on complementary double-stranded DNA.
Moreover, other important variables such as manipulations, clean-up, sample handling, and multi-step procedures comprising each methodological step could affect efficiency and yield, and they should be considered as well. By this manner, a multivariable system could be contemplated that could reveal additional factors on the road ahead in genetics and genomics research [24].
To find new solutions to the needs and challenges described, a multidisciplinary research field should be opened up. However, the control of the nanoscale and nanotechnology production could lead to new approaches and proofs of concepts with potential transfer to NGS technology.
Enzymatic machineries can be used to amplify genomic material such as the well-known polymerase chain reaction (PCR). For example, a rapid and efficient DNA isolation method was developed for qPCR-based detection of pathogenic and spoilage bacteria in milk [25] For neophytes, qPCR is a modified PCR-based technology that allows quantifying real-time single-oligonucleotide reading using fluorescent reporter molecules in targeted quantifications [26].
Robust technology has been developed based on natural complex enzymatic machineries such as PCR. Recently, a gene-based precision medicine technology known as clustered regularly interspaced short palindromic repeats (CRISPR) was created [27]. This biotechnology was developed to repair genomic material by incorporating new oligonucleotide sequences [28]. This is based on a complex enzymatic system that acts as an enzymatic scissor in targeted DNA sequences coupled to the replacement of genomic material [29]. Recently, two researchers working at Max Planck Unit for the Science of Pathogens, Germany, and at University of California, USA, were awarded the Nobel Prize in Chemistry 2020 for “the development of a method for genome editing” [30]. Therefore, CRISPR/Cas9 is still being studied to modify the DNA of animals, plants, and micro-organisms with extremely high precision. This technique, which is able to control fragmentation and re-incorporation of new genomic material, is particularly interesting when transferred to new strategies for DNA detection. Thus, CRISPR could generate different DNA-detection systems by adding fluorescence labelers [31][32].
From the chemical modification of nucleotides and DNA, RNA strands have provided further perspectives by integrating genomics with precision medicine. There are many relevant research studies; however, the combination of a specific nucleotide interactions accompanied by pharmacophores linking and bioconjugations for targeted drug delivery perspectives should be briefly highlighted [33]. Moreover, by this manner, non-covalent interactions are related in the targeted function.
By exploiting the concept of non-classical light from the molecular level by fluorescence labeling such as DNA-intercalating agents or by covalent bond modifications, the research work for new imaging-based developments should be continued. Recently, research has shown an integrated imaging and computational strategy to model gene folding with nucleosome resolution [34]. It was thus possible to identify a specific distribution of nucleosomes within specific genes in super resolution through the simultaneous visualization of DNA and histones. This method advanced information on chromatin accessibility for regulatory factors such as RNA polymerase II. Intercellular variability, transcriptional-dependent gene conformation, and folding of housekeeping and pluripotency-related genes were studied in human pluripotent and differentiated cells, gathering accurate data.
To develop further resolution to nano-biostructures, the molecular level, from the bottom-up design that could provide targeted biomolecular detection from proteins to amino acids and small molecules, should be evaluated. Similarly, the resolution of single nucleotides could prove a major development; yet, this level of resolution is only achieved by a highly smart responsive strategy. In this context, biophotonic strategies for single-molecule detection (SMD) level should be considered [35]. Therefore, there is a growing body of literature on DNA detection and amplification based on different optical approaches and controlled DNA grafting of surfaces [36]. For single-molecule sequencing, researchers may refer to the sequencing and tracking of individual nucleotides based on templated DNA exposed to a solution containing DNA polymerase and a fluorescent nucleotide. If a nucleotide were incorporated, it would be achieved by a complementary strand of the template. Thus, the fluorescence would be read using a total internal reflection fluorescence (TIRF)-based helioscope, recording the positions where the DNA strand had incorporated a fluorescent nucleotide from the solution [37]. It should also be noted that real-time DNA sequencing from single polymerase molecules [38] enabled tracking of single-nucleotide incorporation in real time by fluorescence imaging. The level of developments achieved in the single-molecule dynamic detection of chemical reactions [39] based on an electrochemical device is also worth noting.
Previous developments anticipated the beginning of many challenges to overcome, where biostructure and enzymes coupled to fluorescence labeling joined aptamer interactions with targeted recognitions of specific sequences, or single nucleotides were joined to optimize genomic detection. Results showed high performance; however, the key variable of low genetic material in real samples was managed by amplification. This methodology is linked to a costly and time-consuming process. Therefore, PCR-free strategies are still being developed, specifically in NGS technology.
Nanoscale control has led to new strategies to overcome low DNA concentrations. This is due to many reasons, mainly: (i) proximity of the nanoscale to the molecular level; (ii) molecular hinderance; (iii) interactions associated with non-covalent binding and physical phenomena; and (iv) amplification of the signal such as laser-based materials by different enhanced physical and chemical pathways under study.
A PCR-free DNA-detection strategy was developed from modified magnetic beads with short, targeted DNA aptamers incorporated in microelectromagnetic traps [40]. This enabled the interaction of targeted and complementary DNA strands using a fluorescent polymeric transducer in a miniaturized device that leaded to the separation of the genomic material and detection by laser fluorescence microscopy. Thus, a detection limit of ∼200 target copies in a probed volume of 150 µL (1.4 copies/µL) was obtained for a DNA sequence specific to Candida albicans. Researchers should note that this detection strategy does not require the release of the hybridized target DNA prior to its detection, labeling, or amplification of nucleic acids. The preconcentration and detection steps could be achieved simultaneously on the same support. The design could be extended to the application of this ultrasensitive biosensor to biological samples with complex matrixes and to integrated lab-on-a-chip platforms, where faster multitarget DNA detections could be developed in point-of-care diagnostics and field analysis. The application of in-flow methodologies could also provide other ways of addressing this challenge such as in real-time monitoring of bead-based DNA hybridization in a microfluidic system and the study of amplicon hybridization behavior on solid supports [41].
This development and device-based technology used for fundamental and applied research has provided interesting insights. For instance, the length of the overhanging (dangling) end of target DNA strands following hybridization to the capture probe was correlated with interactions with the complementary strand in solution, which could result in the unbinding of the target and its release from the surface. Thus, an instrument for real-time monitoring of DNA hybridization was created on spherical particles functionalized with oligonucleotide capture probes and arranged in the form of a tightly packed monolayer bead bed inside a microfluidic cartridge. This approach allowed further elucidation of the dynamics of interaction. Thus, it was known that by increasing the length of the dangling end, desorption of target amplicons from bead-bound capture probes is produced at a rate approaching that of the initial hybridization process. This led to the optimization of conditions in order to perform hybridization events from Streptococcus agalactiae cfb gene amplicons obtained from randomized clinical samples. The methodology presented was also used for further investigation of competitive hybridization mechanisms on solid supports to evaluate suitability, such as microarray capture probes. Similarly, other types of nanodevices, microdevices, in-flow devices, and waveguides could be designed [42].
By incorporating plasmonic nanomaterials with an exactly controlled shape and size of each of these parts and placed by controlled 2D patterns, it was possible to tune surface plasmon polaritons (SPPs) arising from metal/semiconductor interfaces [43], enhancing light emission in light-emitting devices [44]. Then, the most recent studies on optical surfaces by surface nanopatterning based on plasmonic resonance allowed for controlling the polarization of light [45], which led to on-chip applications based on six metasurface arrays [46]. In addition, interest in these subwavelength polarizations of light will lead to developments associated with the efficiency of organic light-emitting diodes (OLEDs) by controlling the orientation of the transition dipole moment of the emitters and the resonance of the nanoarray resonators [47]. Then, from the combination of semiconductors and plasmonic nanomaterials, studies into luminescence energy transfer, such as ZnO platforms for enhanced directional fluorescence applications, have been conducted [48] and are still in progress.
In addition, research on semiconductors and nanomaterials incorporated in silica waveguides could develop light guides with atomically thin materials for on-chip near-field plasmon detection by semiconductors such as molybdenum disulfide [49]. In this context, the sensitivity of plasmonic waves and the generation of photons from luminescent phenomena are currently being studied. As for example, filtering the time-resolved luminescence signal of quantum optical circuits by single excitation of well-ordered InGaAs quantum dots generated fluorescence resonance at the nanometer scale [50]. Moreover, novel approaches into modified silica waveguides by polymeric stamps were devised to produce several microns in length of one-dimensional nanoparticle assemblies. Combining this technique with the use of tunable, metaldielectric, core-shell nanoparticles [51] allowed obtaining linear assemblies with adjustable inter-particle separation for luminescence propagation applications within resonant plasmonic waveguides [52]. Therefore, the ultraluminescence generation in silica substrates from Metal Enhanced Fluorescence phenomena (MEF) for chemical sensing to be applied in on-chip [53] and in-flow applications are increasing interest in biosensing [54]. The ultraluminescence properties based on MEF by interaction of the electromagnetic field in the near field of the plasmon with fluorophores can be explained by different pathways such as (i) increased absorption with a higher upper excited state occupation; (ii) decreased non-radiative decays; and (iii) increased radiative decay. Therefore, this effect depends on the distance of the fluorophore from the metallic surface since the electromagnetic field intensity decays exponentially (1/r6), drastically affecting fluorophore excitation [55].
To conclude, insights gained from nanomaterials applied to biosensing can give rise to new approaches for targeted studies in NGS, and in this context, it should be highlighted as an example how the development of better control of materials and modified substrates with nanopores has led to their incorporation in real technology on the market. For example, nanopore genome sequencers [56] based on the DNA strands passing through biological pores permitted their identification. The detection was by measuring the difference in their electrical conductivity while the DNA passes through the pores. This level of technology has recently shown interesting applications at the level of rapid identification of pathogens, antibiotic resistance genes, and plasmids in blood cultures in real time [57]. Extracted bacterial DNA of positive blood cultures led to a comprehensive analysis for diagnostic purposes with an acceptable degree of accuracy for these types of samples. Developments from this mentioned study have opened up new possibilities for future applications in clinical microbiology, healthcare surveillance, and precision medicine.
Finally, in this context, it should be highlighted how nanotechnology is developed within the close relationship between the sizes and physical phenomena involved in the genomic materials, leading to a focus on the nanoscale accompanied by advanced optical setups. Therefore, how the spectroscopy is being developed should be highlighted along with the fact that new spectroscopical techniques are progressively showing insights into biodetection and related research fields and applications, for example, MEF-based strategies, surface enhanced Raman spectroscopy (SERS), and related techniques. In this manner, the engineering of new optical setups is still in progress, improving techniques and methods by incorporating colloidal dispersions of optical active nanomaterials as well as modified substrates to prepare optical active surfaces and porous materials, too [58]. This concept opens a broad vision to develop microarrays towards nanoarrays and NGS [59]. Thus, it is up to a multidisciplinary research point of view to propose new materials and technology associated with NGS and biosensing.
This entry is adapted from the peer-reviewed paper 10.3390/bios13020260
References
- Hermanson, G.T. Bioconjugate Techniques, 2nd ed.; Pierce Biotechnology, ThermoFischer Scientific: Rockford, IL, USA, 2008.
- Nguyen, H.L.; Nam Nguyen, H.; Hai Nguyen, H.; Quynh Luu, M.; Hieu Nguyen, M. Nanoparticles: Synthesis and applications in life science and environmental technology. Adv. Nat. Sci. Nanosci. Nanotechnol. 2015, 6, 015008.
- Hodson, R. Precision Medicine. Nature 2016, 537, s49.
- Zhang, J.; Zhang, T.; Gao, J. Biocompatible Iron Oxide Nanoparticles for Targeted Cancer Gene Therapy: A Review. Nanomaterials 2022, 12, 3323.
- He, W.; Ma, G.; Shen, Q.; Tang, Z. Engineering Gold Nanostructures for Cancer Treatment: Spherical Nanoparticles, Nanorods, and Atomically Precise Nanoclusters. Nanomaterials 2022, 12, 1738.
- Wang, H.; Liu, Y.; Chen, Z.; Sun, L.; Zhao, Y. Anisotropic structural color particles from colloidal phase separation. Sci. Adv. 2020, 6, eaay1438.
- Akbay, N.; Ray, K.; Chowdhury, M.H.; Lakowicz, J.R. Plasmon-controlled fluorescence and single DNA strand sequenching. Proc. SPIE 2012, 8234, 82340M.
- Yu, Y.; Xiao, T.-H.; Wu, Y.; Li, W.; Zeng, Q.-G.; Long, L.; Lic, Z.-Y. Roadmap for single-molecule surface-enhanced Raman spectroscopy. Adv. Photonics 2020, 2, 014002–014020.
- Gomez Palacios, L.R.; Bracamonte, A.G. Development of Nano-, Microdevices for the next generation of Biotechnology, Wearables and miniaturized Instrumentation. RSC Adv. 2022, 12, 12806–12822.
- Bracamonte, A.G. Gold nanoparticles chemical surface modifications as versatile Nanoplatform strategy for fundamental Research towards Nanotechnology and further applications. Nanosci. Nanotechnol. Open Access 2022, 1, 1004.
- Habuchi, S.; Ando, R.; Dedecker, P.; Verheijen, W.; Mizuno, H.; Miyawaki, A.; Hofkens, J. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. PNAS 2005, 102, 9511–9516.
- Nair, R.V.; Jian Yi, P.; Padmanabhan, P.; Gulyas, B.; Murukeshan, V.M. Au nano-urchins enabled localized surface plasmon resonance sensing of beta amyloid Fibrillation. Nanoscale Adv. 2020, 2, 2693–2698.
- Rajil, N.; Sokolov, A.; Yi, Z.; Adams, G.; Agarwal, G.; Belousov, V.; Brick, R.; Chapin, K.; Cirillo, J.; Deckert, V.; et al. A fiber optic–nanophotonic approach to the detection of antibodies and viral particles of COVID-19. Nanophotonics 2021, 10, 235–246.
- Demming, A. DNA sequencing: Nanotechnology unravels the code for life. Nanotechnol. Editor. Lett. 2015, 26, 310201.
- Voelkerding, K.V.; Dames, S.A.; Durtschi, J.D. Next-generation sequencing: From basic research to diagnostics. Clin. Chem. 2009, 55, 641–658.
- Gomez Palacios, L.R.; Bracamonte, A.G. Generation of Bioimaging towards design of hybrid micro-machines and micro-swimmers. J. Chem. Res. Adv. JCRA 2022, 3, 22–27.
- Gomez Palacios, L.R.; Martinez, S.; Tettamanti, C.; Quinteros, D.; Bracamonte, A.G. Nano-chemistry and Bio-conjugation with perspectives on the design of Nano-Immune platforms, vaccines and new combinatorial treatments. J. Vaccines Immunol. 2021, 7, 049–056.
- Børresen, A.-L. Mismatch detection using heteroduplex analysis. Chapter 7: Unit 7.3. Curr. Protoc. Hum. Genet 2002, 1, 1–25.
- Taberlet, P.; Griffin, S.; Goossens, B.; Questiau, S.; Manceau, V.; Escaravage, N.; Waits, L.P.; Bouvet, J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996, 24, 3189–3194.
- Li, N.; Ho, C.-M. Aptamer-Based Optical Probes with Separated Molecular Recognition and Signal Transduction Modules. J. Am. Chem. Soc. 2008, 130, 2380–2381.
- Menking, D.; Emanuel, P.A.; Valdes, P.P.; Kracke, S.K. Rapid cleanup of bacterial DNA from field samples. Resour. Conserv. Recycl. 1999, 27, 179–186.
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Isolating, Cloning, and Sequencing DNA, Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002.
- Next-Generation Sequencing (NGS). Explore the Technology. Available online: https://www.illumina.com/science/technology/next-generation-sequencing.html (accessed on 20 January 2023).
- McGuire, A.L.; Gabriel, S.; Tishkoff, S.A.; Wonkam, A.; Chakravarti, A.; Furlong, E.E.M.; Treutlein, B.; Meissner, A.; Chang, H.Y.; López-Bigas, N.; et al. The road ahead in genetics and genomics. Nat. Rev. Genet. 2020, 21, 581–596.
- Pancza, B.; Szathmáry, M.; Gyurján, I.; Bánkuti, B.; Tudós, Z.; Szathmary, S.; Zsófia, L.S.; Sipos-Kozma, Á.B.; Varga, L.; Szenthe, K.; et al. A rapid and efficient DNA isolation method for qPCR-based detection of pathogenic and spoilage bacteria in milk. Food Control 2021, 130, 108236.
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622.
- Kararoudi, M.N.; Hejazi, S.S.; Elmas, E.; Hellström, M.; Naeimi Kararoudi, M.; Padma, A.M.; Lee, D.; Dolatshad, H. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Gene Editing Technique in Xenotransplantation. Front. Immunol. 2018, 9, 1–7.
- Kan, Y.; Ruis, B.; Takasugi, T.; Hendrickson, E.A. Mechanisms of precise genome editing using oligonucleotide donors. Genome Res. 2017, 7, 1099–1111.
- Burgess, D.J. A CRISPR genome-editing tool. Nat. Rev. Genet. 2013, 81, 1–2.
- Charpentier, E.J.; Doudna, A. Popular Science Background. The Nobel Prize in Chemistry 2020. “Genetic Scissors: A Tool for Rewriting the Code of Life”. Royal Swedish Academy of Sciences: Stockholm, Sweden, 2020; pp. 1–8.
- Osborn, M.J.; Bhardway, A.; Bingea, S.P.; Knipping, F.; Feser, C.J.; Lees, C.J.; Collins, D.P.; Steer, C.J.; Blazar, B.R.; Tolar, J. CRISPR/Cas9-Based Lateral Flow and Fluorescence Diagnostics. Bioengineering 2021, 8, 23.
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2022, 5, 643–656.
- Campagne, S.; Boigner, S.; Rüdisser, S.; Moursy, A.; Gillioz, L.; Knörlein, A.; Hall, J.; Ratni, H.; Cléry, A.; Allain, F. H-T. Structural basis of a small molecule targeting RNA for a specific splicing correction. Nat. Chem. Biol. 2019, 15, 1191–1198.
- Neguembor, M.V.; Arcon, J.P.; Buitrago, D.; Lema, R.; Walther, J.; Garate, X.; Martin, L.; Romero, P.; Al Haj Abed, J.; Gut, M.; et al. MiOS, an integrated imaging and computational strategy to model gene folding with nucleosome resolution. Nat. Struct. Mol. Biol. 2022, 29, 1011–1023.
- Bracamonte, A.G. Biophotonics at Single-Molecule Detection Level. In Frontiers in Nano- and Micro-Device Design for Applied Nanophotonics, Biophotonics and Nanomedicine; Bentham Science Publishers: Sharjah, United Arab Emirates, 2021; Chapter 11; ISBN 978-1-68108-857-0.
- Walt, D.R. Optical Methods for Single Molecule Detection and Analysis. Anal. Chem. 2013, 85, 1258–1263.
- Braslavsky, I.; Hebert, B.E.; Kartalov, S.R. Sequence information can be obtained from single DNA molecules. Proc. Natl. Acad. Sci. USA 2003, 100, 3960−3964.
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-Time DNA Sequencing from Single Polymerase. Mol. Sci. 2009, 323, 133–138.
- Guan, J.; Jia, C.; Li, Y.; Liu, Z.; Wang, J.; Yang, Z.; Gu, C.; Su, D.; Houk, K.N.; Zhang, D.; et al. Direct single molecule dynamic detection of chemical reactions. Sci. Adv. 2018, 4, eaar2177.
- Dubus, S.; Gravel, J.-F.; Le Drogoff, B.; Nobert, P.; Veres, T.; Boudreau, D. PCR-Free DNA Detection Using a Magnetic Bead-Supported Polymeric Transducer and Microelectromagnetic Traps. Anal. Chem. 2006, 78, 4457–4464.
- Boissinot, K.; Peytavi, R.; Chapdelaine, S.; Geissler, M.; Boissinot, M.; Martel, E.A.; Béliveau-Viel, D.; Gravel, J.-F.; Malic, L.; Veres, T.; et al. Real-time monitoring of bead-based DNA hybridization in a microfluidic system: Study of amplicon hybridization behavior on solid supports. Analyst 2021, 146, 4226–4234.
- Salinas, C.; Bracamonte, A.G. From Microfluidics to Nanofluidics and signal Wave-guiding for Nanophotonics, Biophotonics resolution and Drug Delivery. Front. Drug Chem. Clin. Res. 2019, 2, 1–6.
- Barnes, W.L. Electromagnetic crystals for surface plasmon polaritons and the extraction of light from emissive devices. J. Light. Tech. 1999, 17, 2170–2182.
- Kim, J.Y.; Kwon, M.K.; Lee, K.S.; Park, S.J.; Kim, S.H.; Lee, K.D. Enhanced light extraction from GaN-based green light-emitting diode with photonic crystal. Appl. Phys. Lett. 2007, 91, 181109.
- Chen, W.; Tymchenko, M.; Gopalan, P.; Ye, X.; Wu, Y.; Zhang, M.; Murra, C.B.; Alu, A.; Kagan, C.R. Large-Area Nanoimprinted Colloidal Au Nanocrystal-Based Nanoantennas for Ultrathin Polarizing Plasmonic Metasurfaces. Nano Lett. 2015, 15, 5254–5260.
- Wu, P.C.; Tsai, W.Y.; Ti Chen, Y.W.; Huang, T.Y.; Chen, J.-W.; Chen, C.Y.; Lia, C.H.; Chu, T.; Sun, G.; Tsai, D.P. Integrated metasurface chip for versatile polarization generation. SPIE Newsroom 2017, 1–3.
- Senes, A.; Meskers, S.C.J.; Greiner, H.; Suzuki, K.; Kaji, H.; Adachi, C.; Wilsona, J.S.; Janssen, R.A.J. Increasing the horizontal orientation of transition dipole moments in solution processed small molecular emitters. J. Mater. Chem. C 2017, 5, 6555–6562.
- Lai, C.W.; An, J.; Ong, H.C. Surface-plasmon-mediated emission from metal-capped ZnO thin films. Appl. Phys. Lett. 2005, 86, 251105.
- Goodfellow, K.M.; Chakraborty, C.; Beams, R.; Novotny, L.; Nick Vamivakas, A. Direct On-Chip Optical Plasmon Detection with an Atomically Thin Semiconductor. Nano Lett. 2015, 15, 5477–5481.
- Reithmaier, G.; Kaniber, M.; Flassig, F.; Lichtmannecker, S.; Müller, K.; Andrejew, A.; Vučković, J.; Gross, R.; Finley, J.J. On-Chip Generation, Routing, and Detection of Resonance Fluorescence. Nano Lett. 2015, 15, 5208–5213.
- Asselin, J.; Legros, P.; Grégoire, A.; Boudreau, D. Correlating metal-enhanced 2 core-shell nanoparticles. Plasmonics 2016, 5, 1–8.
- Grégoire, A.; Boudreau, D. Metal-Enhanced Fluorescence in Plasmonic Waveguides. In Nano-Optics: Principles Enabling Basic Research and Applications; Springer Science + Business Media: Dordrecht, The Netherlands, 2017; Chapter 28.
- Yen, C.-W.; de Puig, H.; Tam, J.O.; Gómez-Márquez, J.; Bosch, I.; Hamad-Schifferli, K.; Gehrk, L. Multicolored silver nanoparticles for multiplexed disease diagnostics: Distinguishing dengue, yellow fever, and Ebola viruses. Lab Chip. 2015, 15, 1638–1640.
- Glass, N.R.; Tjeung, R.; Chan, P.; Yeo, L.Y.; Friend, J.R. Organosilane deposition for microfluidic application. Biomicrofluidics 2011, 5, 036501.
- Lackowicz, J.R. Radiative decay engineering: Metal enhanced fluorescence and plasmon emission. Anal. Biochem. 2005, 337, 171–194.
- Bowden, R. Sequencing of human genomes with nanopore technology. Nat. Commun. 2019, 10, 1869.
- Taxt, A.M.; Avershina, E.; Frye, S.A.; Naseer, U.; Ahmad, R. Rapid identification of pathogens, antibiotic resistance genes and plasmids in blood cultures by nanopore sequencing. Sci. Rep. Springer Nat. 2020, 10, 7622.
- Bracamonte, A.G. Microarrays towards Nanoarrays and the Future Next Generation of Sequencing Methodologies (NGS), Sensing and Bio-Sensing Research; Elsevier: Amsterdam, The Netherlands, 2022; Volume 37, p. 100503.
- Gomez Palacios, L.R.; Veglia, A.; Bracamonte, A.G. Inflow nano-optics from the near-to the far-field detection based on Metal-Enhanced Fluorescence signaling. Microchem. J. 2021, 169, 106539.
This entry is offline, you can click here to edit this entry!
