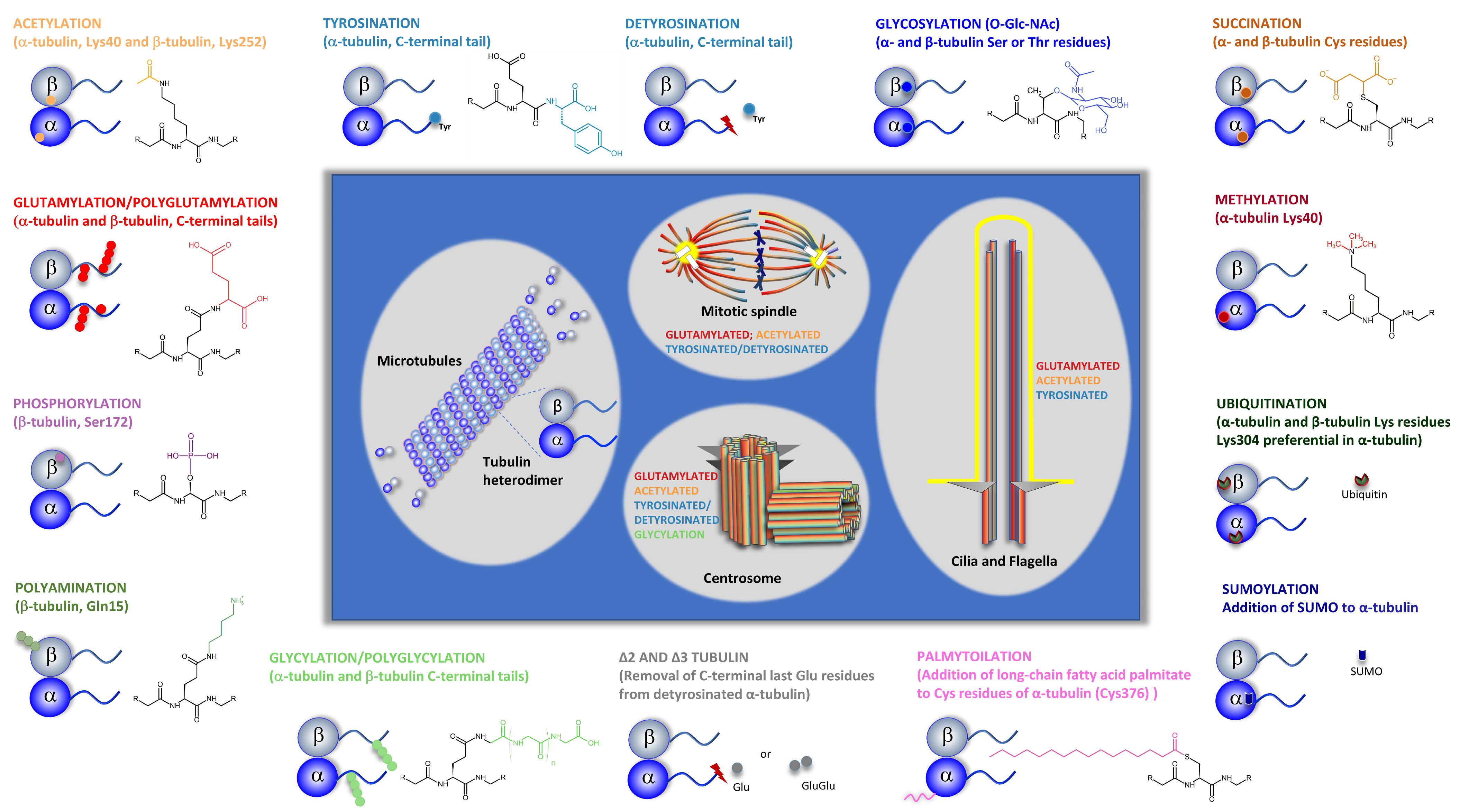
Microtubules (MTs) are dynamic structures that are part of the cell cytoskeleton. They play important roles in various cellular functions, such as intracellular organization and transport, cell division, and cell migration. MTs are made up of α/β-tubulin heterodimers that display diversity due to the existence of different tubilin isotypes and post-translational modifications (PTMs). One specific PTM, tubulin-acetylation, occurs inside the MT lumen and has been pointed out as a hallmark of stable old MTs. However, the question if it is a cause or a consequence of long-lived MTs has never been clarified. The view on tubulin acetylation is that this modification alters the mechanical properties of MTs allowing MTs to bend and to resist age-related lattice damage caused by multiple interactions with different factors during their existence. However, how this ability of MTs to survive structural damage is translated into specific cellular functions is still controversial, and it is far from being elucidated. This PTM is also associated with cellular responses to stress and various human pathologies.The regulation of enzymes involved in tubulin acetylation and deacetylation is important for maintaining proper cell physiology.
- acetylation
- tubulin
- Lys40
- microtubules
- post-translational modifications
- αTAT1
- HDAC6
- SIRT2
- microtubule-associated proteins
- microtubule-mechanical properties
1. Introduction
2. The Dynamic Nature of Microtubules
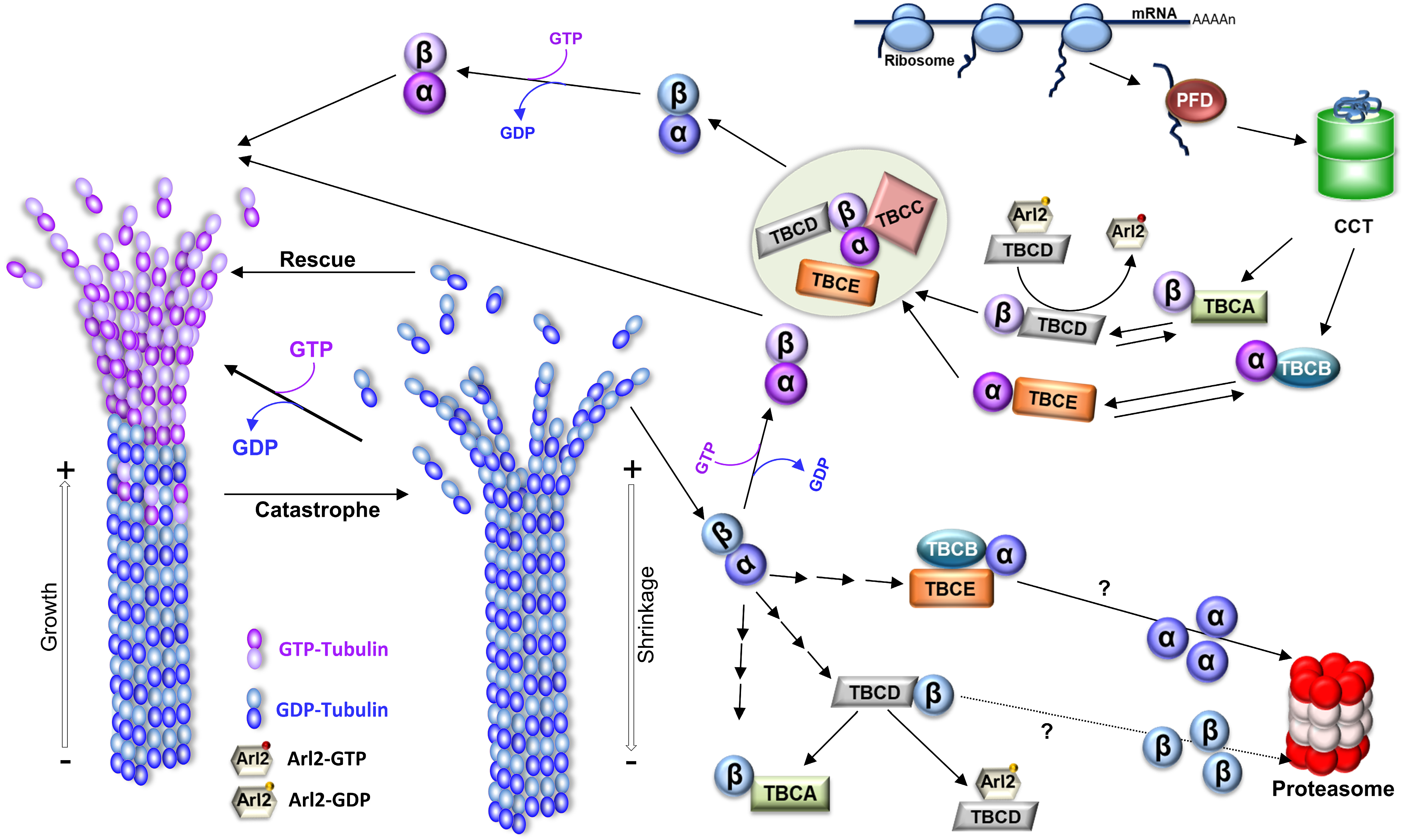
3. The Functional Diversity of Microtubules and Microtubule-Based Structures
4. In Vivo Microtubule Dynamics Is Modulated by Microtubule-Associated Proteins
5. The In Vivo Diversity of Tubulin Pools and Microtubule Functional Diversity
This entry is adapted from the peer-reviewed paper 10.3390/biology12040561
References
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577.
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580.
- Wloga, D.; Joachimiak, E.; Fabczak, H. Tubulin Post-Translational Modifications and Microtubule Dynamics. Int. J. Mol. Sci. 2017, 18, 2207.
- Gudimchuk, N.B.; McIntosh, J.R. Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol. 2021, 22, 777–795.
- Gonçalves, J.; Tavares, A.; Carvalhal, S.; Soares, H. Revisiting the tubulin folding pathway: New roles in centrosomes and cilia. Biomol. Concepts 2010, 1, 423–434.
- Cowan, N.J.; Lewis, S.A. Type II chaperonns, prefoldin, and the tubulin-specific chaperones. Adv. Protein Chem. 2001, 59, 73–104.
- Lopez, T.; Dalton, K.; Frydman, J. The Mechanism and Function of Group II Chaperonins. J. Mol. Biol. 2015, 427, 2919–2930.
- Lewis, S.A.; Tian, G.; Cowan, N.J. The α- and β-tubulin folding pathways. Trends Cell Biol. 1997, 7, 479–484.
- Lopez-Fanarraga, M.; Avila, J.; Guasch, A.; Coll, M.; Zabala, J.C. Review: Postchaperonin Tubulin Folding Cofactors and Their Role in Microtubule Dynamics. J. Struct. Biol. 2001, 135, 219–229.
- Kortazar, D.; Fanarraga, M.; Carranza, G.; Bellido, J.; Villegas, J.; Avila, J.; Zabala, J. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp. Cell Res. 2007, 313, 425–436.
- Voloshin, O.; Gocheva, Y.; Gutnick, M.; Movshovich, N.; Bakhrat, A.; Baranes-Bachar, K.; Bar-Zvi, D.; Parvari, R.; Gheber, L.; Raveh, D. Tubulin chaperone E binds microtubules and proteasomes and protects against misfolded protein stress. Cell. Mol. Life Sci. 2010, 67, 2025–2038.
- Desai, A.; Mitchison, T.J. Microtubule Polymerization Dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117.
- Serna, M.; Zabala, J.C. Tubulin Folding and Degradation. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 1–9.
- Nolasco, S.; Bellido, J.; Serna, M.; Carmona, B.; Soares, H.; Zabala, J.C. Colchicine Blocks Tubulin Heterodimer Recycling by Tubulin Cofactors TBCA, TBCB, and TBCE. Front. Cell Dev. Biol. 2021, 9, 950.
- Roll-Mecak, A. The Tubulin Code in Microtubule Dynamics and Information Encoding. Dev. Cell 2020, 54, 7–20.
- Camelo, C.; Peneda, C.; Carmona, B.; Soarets, H. TBCC. In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer: New York, NY, USA, 2016; pp. 1–8.
- Lomakin, A.J.; Semenova, I.; Zaliapin, I.; Kraikivski, P.; Nadezhdina, E.; Slepchenko, B.M.; Akhmanova, A.; Rodionov, V. CLIP-170-Dependent Capture of Membrane Organelles by Microtubules Initiates Minus-End Directed Transport. Dev. Cell 2009, 17, 323–333.
- Lomakin, A.J.; Kraikivski, P.; Semenova, I.; Ikeda, K.; Zaliapin, I.; Tirnauer, J.S.; Akhmanova, A.; Rodionov, V. Stimulation of the CLIP-170–dependent capture of membrane organelles by microtubules through fine tuning of microtubule assembly dynamics. Mol. Biol. Cell 2011, 22, 4029–4037.
- Kanfer, G.; Peterka, M.; Arzhanik, V.K.; Drobyshev, A.L.; Ataullakhanov, F.I.; Volkov, V.A.; Kornmann, B. CENP-F couples cargo to growing and shortening microtubule ends. Mol. Biol. Cell 2017, 28, 2400–2409.
- Seetharaman, S.; Etienne-Manneville, S. Microtubules at focal adhesions—A double-edged sword. J. Cell Sci. 2019, 132, jcs232843.
- Stehbens, S.; Wittmann, T. Targeting and transport: How microtubules control focal adhesion dynamics. J. Cell Biol. 2012, 198, 481–489.
- Stehbens, S.J.; Paszek, M.; Pemble, H.; Ettinger, A.; Gierke, S.; Wittmann, T. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nature 2014, 16, 558–570.
- Kopf, A.; Kiermaier, E. Dynamic Microtubule Arrays in Leukocytes and Their Role in Cell Migration and Immune Synapse Formation. Front. Cell Dev. Biol. 2021, 9, 635511.
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735.
- Mayor, R.; Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016, 17, 97–109.
- Bayless, K.J.; Johnson, G.A. Role of the Cytoskeleton in Formation and Maintenance of Angiogenic Sprouts. J. Vasc. Res. 2011, 48, 369–385.
- Hohmann, T.; Dehghani, F. The Cytoskeleton—A Complex Interacting Meshwork. Cells 2019, 8, 362.
- Bornens, M. Organelle positioning and cell polarity. Nat. Rev. Mol. Cell Biol. 2008, 9, 874–886.
- Budovsky, A.; Fraifeld, V.E.; Aronov, S. Linking cell polarity, aging and rejuvenation. Biogerontology 2010, 12, 167–175.
- Soares, H.; Marinho, H.S.; Real, C.; Antunes, F. Cellular polarity in aging: Role of redox regulation and nutrition. Genes Nutr. 2013, 9, 371.
- Vasileva, E.; Citi, S. The role of microtubules in the regulation of epithelial junctions. Tissue Barriers 2018, 6, 1–20.
- Matis, M. The Mechanical Role of Microtubules in Tissue Remodeling. Bioessays 2020, 42, e1900244.
- Singh, A.; Saha, T.; Begemann, I.; Ricker, A.; Nüsse, H.; Thorn-Seshold, O.; Klingauf, J.; Galic, M.; Matis, M. Polarized microtubule dynamics directs cell mechanics and coordinates forces during epithelial morphogenesis. Nat. Cell Biol. 2018, 20, 1126–1133.
- Ghalloussi, D.; Dhenge, A.; Bergmeier, W. New insights into cytoskeletal remodeling during platelet production. J. Thromb. Haemost. 2019, 17, 1430–1439.
- Patel, S.R.; Richardson, J.L.; Schulze, H.; Kahle, E.; Galjart, N.; Drabek, K.; Shivdasani, R.A.; Hartwig, J.H.; Italiano, J.J.E. Differential roles of microtubule assembly and sliding in proplatelet formation by megakaryocytes. Blood 2005, 106, 4076–4085.
- Thon, J.N.; Macleod, H.; Begonja, A.J.; Zhu, J.; Lee, K.-C.; Mogilner, A.; Hartwig, J.H.; Jr, J.E.I. Microtubule and cortical forces determine platelet size during vascular platelet production. Nat. Commun. 2012, 3, 852.
- Sánchez, I.; Dynlacht, B.D. Cilium assembly and disassembly. Nat. Cell Biol. 2016, 18, 711–717.
- Pitaval, A.; Senger, F.; Letort, G.; Gidrol, X.; Guyon, L.; Sillibourne, J.; Théry, M. Microtubule stabilization drives 3D centrosome migration to initiate primary ciliogenesis. J. Cell Biol. 2017, 216, 3713–3728.
- van der Vaart, B.; Akhmanova, A.; Straube, A. Regulation of microtubule dynamic instability. Biochem. Soc. Trans. 2009, 37, 1007–1013.
- Bodakuntla, S.; Jijumon, A.; Villablanca, C.; Gonzalez-Billault, C.; Janke, C. Microtubule-Associated Proteins: Structuring the Cytoskeleton. Trends Cell Biol. 2019, 29, 804–819.
- Kapoor, T.M. Metaphase Spindle Assembly. Biology 2017, 6, 8.
- Starr, D.A. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 2009, 122, 577–586.
- Dogterom, M.; Kerssemakers, J.W.J.; Romet-Lemonne, G.; E Janson, M. Force generation by dynamic microtubules. Curr. Opin. Cell Biol. 2005, 17, 67–74.
- Kimura, A.; Onami, S. Modeling Microtubule-Mediated Forces and Centrosome Positioning in Caenorhabditis elegans Embryos. Methods Cell Biol. 2010, 97, 437–453.
- Burakov, A.; Nadezhdina, E.; Slepchenko, B.; Rodionov, V. Centrosome positioning in interphase cells. J. Cell Biol. 2003, 162, 963–969.
- Zhu, J.; Burakov, A.; Rodionov, V.; Mogilner, A. Finding the Cell Center by a Balance of Dynein and Myosin Pulling and Microtubule Pushing: A Computational Study. Mol. Biol. Cell 2010, 21, 4418–4427.
- Wu, J.; Misra, G.; Russell, R.J.; Ladd, A.J.C.; Lele, T.P.; Dickinson, R.B. Effects of dynein on microtubule mechanics and centrosome positioning. Mol. Biol. Cell 2011, 22, 4834–4841.
- Tanimoto, H.; Sallé, J.; Dodin, L.; Minc, N. Physical forces determining the persistency and centring precision of microtubule asters. Nat. Phys. 2018, 14, 848–854.
- Odell, J.; Sikirzhytski, V.; Tikhonenko, I.; Cobani, S.; Khodjakov, A.; Koonce, M. Force balances between interphase centrosomes as revealed by laser ablation. Mol. Biol. Cell 2019, 30, 1705–1715.
- Daga, R.R.; Yonetani, A.; Chang, F. Asymmetric Microtubule Pushing Forces in Nuclear Centering. Curr. Biol. 2006, 16, 1544–1550.
- Tran, P.; Marsh, L.; Doye, V.; Inoué, S.; Chang, F. A Mechanism for Nuclear Positioning in Fission Yeast Based on Microtubule Pushing. J. Cell Biol. 2001, 153, 397–412.
- Zhao, T.; Graham, O.S.; Raposo, A.; Johnston, D.S. Growing Microtubules Push the Oocyte Nucleus to Polarize the Drosophila Dorsal-Ventral Axis. Science 2012, 336, 999–1003.
- Letort, G.; Nedelec, F.; Blanchoin, L.; Théry, M. Centrosome centering and decentering by microtubule network rearrangement. Mol. Biol. Cell 2016, 27, 2833–2843.
- Théry, M.; Racine, V.; Piel, M.; Pépin, A.; Dimitrov, A.; Chen, Y.; Sibarita, J.-B.; Bornens, M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA 2006, 103, 19771–19776.
- Komarova, Y.; De Groot, C.O.; Grigoriev, I.; Gouveia, S.M.; Munteanu, E.L.; Schober, J.M.; Honnappa, S.; Buey, R.M.; Hoogenraad, C.C.; Dogterom, M.; et al. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 2009, 184, 691–706.
- Akhmanova, A.; Steinmetz, M.O. Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008, 9, 309–322.
- Lansbergen, G.; Akhmanova, A. Microtubule Plus End: A Hub of Cellular Activities. Traffic 2006, 7, 499–507.
- Honnappa, S.; Gouveia, S.; Weisbrich, A.; Damberger, F.; Bhavesh, N.S.; Jawhari, H.; Grigoriev, I.; van Rijssel, F.J.; Martinez-Buey, R.; Lawera, A.; et al. An EB1-Binding Motif Acts as a Microtubule Tip Localization Signal. Cell 2009, 138, 366–376.
- Vitre, B.; Coquelle, F.M.; Heichette, C.; Garnier, C.; Chrétien, D.; Arnal, I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 2008, 10, 415–421.
- Bieling, P.; Laan, L.; Schek, H.; Munteanu, E.L.; Sandblad, L.; Dogterom, M.; Brunner, D.; Surrey, T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature 2007, 450, 1100–1105.
- Maurer, S.P.; Fourniol, F.J.; Bohner, G.; Moores, C.A.; Surrey, T. EBs Recognize a Nucleotide-Dependent Structural Cap at Growing Microtubule Ends. Cell 2012, 149, 371–382.
- Seetapun, D.; Castle, B.T.; McIntyre, A.J.; Tran, P.T.; Odde, D.J. Estimating the Microtubule GTP Cap Size In Vivo. Curr. Biol. 2012, 22, 1681–1687.
- Zhang, R.; Alushin, G.M.; Brown, A.; Nogales, E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell 2015, 162, 849–859.
- Duellberg, C.; Cade, N.I.; Holmes, D.; Surrey, T. The size of the EB cap determines instantaneous microtubule stability. Elife 2016, 5, e13470.
- Kumar, P.; Lyle, K.S.; Gierke, S.; Matov, A.; Danuser, G.; Wittmann, T. GSK3β phosphorylation modulates CLASP–microtubule association and lamella microtubule attachment. J. Cell Biol. 2009, 184, 895–908.
- Lansbergen, G.; Grigoriev, I.; Mimori-Kiyosue, Y.; Ohtsuka, T.; Higa, S.; Kitajima, I.; Demmers, J.; Galjart, N.; Houtsmuller, A.B.; Grosveld, F.; et al. CLASPs Attach Microtubule Plus Ends to the Cell Cortex through a Complex with LL5β. Dev. Cell 2006, 11, 21–32.
- Galjart, N. CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, 487–498.
- Walczak, C.E.; Gayek, S.; Ohi, R. Microtubule-Depolymerizing Kinesins. Annu. Rev. Cell Dev. Biol. 2013, 29, 417–441.
- Cassimeris, L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 2002, 14, 18–24.
- Gupta, K.K.; Li, C.; Duan, A.; Alberico, E.O.; Kim, O.V.; Alber, M.S.; Goodson, H.V. Mechanism for the catastrophe-promoting activity of the microtubule destabilizer Op18/stathmin. Proc. Natl. Acad. Sci. USA 2013, 110, 20449–20454.
- Roostalu, J.; Surrey, T. Microtubule nucleation: Beyond the template. Nat. Rev. Mol. Cell Biol. 2017, 18, 702–710.
- McNally, F.J.; Vale, R.D. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 1993, 75, 419–429.
- Evans, K.J.; Gomes, E.; Reisenweber, S.M.; Gundersen, G.G.; Lauring, B.P. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 2005, 168, 599–606.
- Roll-Mecak, A.; Vale, R.D. The Drosophila Homologue of the Hereditary Spastic Paraplegia Protein, Spastin, Severs and Disassembles Microtubules. Curr. Biol. 2005, 15, 650–655.
- Mukherjee, S.; Valencia, J.D.D.; Stewman, S.; Metz, J.; Monnier, S.; Rath, U.; Asenjo, A.B.; Charafeddine, R.A.; Sosa, H.J.; Ross, J.L.; et al. Human Fidgetin is a microtubule severing the enzyme and minus-end depolymerase that regulates mitosis. Cell Cycle 2012, 11, 2359–2366.
- Kuo, Y.-W.; Howard, J. Cutting, Amplifying, and Aligning Microtubules with Severing Enzymes. Trends Cell Biol. 2020, 31, 50–61.
- McNally, F.J.; Roll-Mecak, A. Microtubule-severing enzymes: From cellular functions to molecular mechanism. J. Cell Biol. 2018, 217, 4057–4069.
- Zhang, D.; Grode, K.D.; Stewman, S.F.; Diaz-Valencia, J.D.; Liebling, E.; Rath, U.; Riera, T.; Currie, J.D.; Buster, D.W.; Asenjo, A.B.; et al. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat. Cell Biol. 2011, 13, 361–369.
- E Mains, P.; Kemphues, K.J.; A Sprunger, S.; A Sulston, I.; Wood, W.B. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics 1990, 126, 593–605.
- Ahmad, F.; Yu, W.; McNally, F.J.; Baas, P.W. An Essential Role for Katanin in Severing Microtubules in the Neuron. J. Cell Biol. 1999, 145, 305–315.
- Kapitein, L.C.; Hoogenraad, C.C. Building the Neuronal Microtubule Cytoskeleton. Neuron 2015, 87, 492–506.
- Baas, P.W.; Rao, A.N.; Matamoros, A.J.; Leo, L. Stability properties of neuronal microtubules. Cytoskeleton 2016, 73, 442–460.
- Lohret, T.A.; McNally, F.J.; Quarmby, L.M. A Role for Katanin-mediated Axonemal Severing during Chlamydomonas Deflagellation. Mol. Biol. Cell 1998, 9, 1195–1207.
- Vemu, A.; Szczesna, E.; Zehr, E.A.; Spector, J.O.; Grigorieff, N.; Deaconescu, A.M.; Roll-Mecak, A. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science 2018, 361, eaau1504.
- Komarova, Y.A.; Akhmanova, A.S.; Kojima, S.-I.; Galjart, N.; Borisy, G.G. Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 2002, 159, 589–599.
- Chen, J.; Kanai, Y.; Cowan, N.J.; Hirokawa, N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 1992, 360, 674–677.
- Harada, A.; Oguchi, K.; Okabe, S.; Kuno, J.; Terada, S.; Ohshima, T.; Sato-Yoshitake, R.; Takei, Y.; Noda, T.; Hirokawa, N. Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature 1994, 369, 488–491.
- Shahpasand, K.; Uemura, I.; Saito, T.; Asano, T.; Hata, K.; Shibata, K.; Toyoshima, Y.; Hasegawa, M.; Hisanaga, S.-I. Regulation of Mitochondrial Transport and Inter-Microtubule Spacing by Tau Phosphorylation at the Sites Hyperphosphorylated in Alzheimer’s Disease. J. Neurosci. 2012, 32, 2430–2441.
- Fourniol, F.J.; Sindelar, C.V.; Amigues, B.; Clare, D.K.; Thomas, G.; Perderiset, M.; Francis, F.; Houdusse, A.; Moores, C.A. Template-free 13-protofilament microtubule–MAP assembly visualized at 8 Å resolution. J. Cell Biol. 2010, 191, 463–470.
- Hooikaas, P.J.; Martin, M.; Mühlethaler, T.; Kuijntjes, G.-J.; Peeters, C.A.; Katrukha, E.A.; Ferrari, L.; Stucchi, R.; Verhagen, D.G.; van Riel, W.E.; et al. MAP7 family proteins regulate kinesin-1 recruitment and activation. J. Cell Biol. 2019, 218, 1298–1318.
- Subramanian, R.; Wilson-Kubalek, E.M.; Arthur, C.P.; Bick, M.J.; Campbell, E.A.; Darst, S.A.; Milligan, R.A.; Kapoor, T.M. Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein. Cell 2010, 142, 433–443.
- Bieling, P.; Telley, I.A.; Surrey, T. A Minimal Midzone Protein Module Controls Formation and Length of Antiparallel Microtubule Overlaps. Cell 2010, 142, 420–432.
- Ichikawa, M.; Bui, K.H. Microtubule Inner Proteins: A Meshwork of Luminal Proteins Stabilizing the Doublet Microtubule. Bioessays 2018, 40, 1700209.
- Khalifa, A.A.Z.; Ichikawa, M.; Dai, D.; Kubo, S.; Black, C.S.; Peri, K.; McAlear, T.S.; Veyron, S.; Yang, S.K.; Vargas, J.; et al. The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications. Elife 2020, 9, 52760.
- Ma, M.; Stoyanova, M.; Rademacher, G.; Dutcher, S.K.; Brown, A.; Zhang, R. Structure of the Decorated Ciliary Doublet Microtubule. Cell 2019, 179, 909–922.
- Goodson, H.V.; Jonasson, E.M. Microtubules and Microtubule-Associated Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10, a022608.
- Dumont, E.L.P.; Do, C.; Hess, H. Molecular wear of microtubules propelled by surface-adhered kinesins. Nat. Nanotechnol. 2015, 10, 166–169.
- Triclin, S.; Inoue, D.; Gaillard, J.; Htet, Z.M.; DeSantis, M.E.; Portran, D.; Derivery, E.; Aumeier, C.; Schaedel, L.; John, K.; et al. Self-repair protects microtubules from destruction by molecular motors. Nat. Mater. 2021, 20, 883–891.
- Janke, C. The tubulin code: Molecular components, readout mechanisms, and functions. J. Cell Biol. 2014, 206, 461–472.
- Wloga, D.; Joachimiak, E.; Louka, P.; Gaertig, J. Posttranslational Modifications of Tubulin and Cilia. Cold Spring Harb. Perspect. Biol. 2016, 9, a028159.
- Ludueña, R.F. A Hypothesis on the Origin and Evolution of Tubulin. Int. Rev. Cell Mol. Biol. 2013, 302, 41–185.
- Breuss, M.W.; Leca, I.; Gstrein, T.; Hansen, A.H.; Keays, D.A. Tubulins and brain development—The origins of functional specification. Mol. Cell. Neurosci. 2017, 84, 58–67.
- Kemphues, K.J.; Raff, E.C.; Raff, R.A.; Kaufman, T.C. Mutation in a testis-specific β-tubulin in Drosophila: Analysis of its effects on meiosis and map location of the gene. Cell 1980, 21, 445–451.
- Nielsen, M.G.; Turner, F.; Hutchens, J.A.; Raff, E.C. Axoneme-specific β-tubulin specialization: A Conserved C-Terminal Motif Specifies the Central Pair. Curr. Biol. 2001, 11, 529–533.
- Fukushige, T.; Siddiqui, Z.; Chou, M.; Culotti, J.; Gogonea, C.; Siddiqui, S.; Hamelin, M. MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans. J. Cell Sci. 1999, 112, 395–403.
- Savage, C.; Hamelin, M.; Culotti, J.G.; Coulson, A.; Albertson, D.G.; Chalfie, M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989, 3, 870–881.
- Schwer, H.D.; Lecine, P.; Tiwari, S.; E Italiano, J.; Hartwig, J.H.; A Shivdasani, R. A lineage-restricted and divergent β-tubulin isoform is essential for the biogenesis, structure and function of blood platelets. Curr. Biol. 2001, 11, 579–586.
- Janke, C.; Magiera, M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 307–326.
- Song, Y.; Brady, S.T. Post-translational modifications of tubulin: Pathways to functional diversity of microtubules. Trends Cell Biol. 2014, 25, 125–136.
- Fourest-Lieuvin, A.; Peris, L.; Gache, V.; Garcia-Saez, I.; Juillan-Binard, C.; Lantez, V.; Job, D. Microtubule Regulation in Mitosis: Tubulin Phosphorylation by the Cyclin-dependent Kinase Cdk1. Mol. Biol. Cell 2006, 17, 1041–1050.
- Ori-McKenney, K.M.; McKenney, R.J.; Huang, H.H.; Li, T.; Meltzer, S.; Jan, L.Y.; Vale, R.D.; Wiita, A.P.; Jan, Y.N. Phosphorylation of β-Tubulin by the Down Syndrome Kinase, Minibrain/DYRK1a, Regulates Microtubule Dynamics and Dendrite Morphogenesis. Neuron 2016, 90, 551–563.
- Ludueña, R.F.; Zimmermann, H.-P.; Little, M. Identification of the phosphorylated β-tubulin isotype in differentiated neuroblastoma cells. FEBS Lett. 1988, 230, 142–146.
- Peters, J.D.; Furlong, M.T.; Asai, D.J.; Harrison, M.L.; Geahlen, R.L. Syk, Activated by Cross-linking the B-cell Antigen Receptor, Localizes to the Cytosol Where It Interacts with and Phosphorylates α-Tubulin on Tyrosine. J. Biol. Chem. 1996, 271, 4755–4762.
- Akiyama, T.; Kadowaki, T.; Nishida, E.; Kadooka, T.; Ogawara, H.; Fukami, Y.; Sakai, H.; Takaku, F.; Kasuga, M. Substrate specificities of tyrosine-specific protein kinases toward cytoskeletal proteins in vitro. J. Biol. Chem. 1986, 261, 14797–14803.
- Matten, W.T.; Aubry, M.; West, J.; Maness, P.F. Tubulin is phosphorylated at tyrosine by pp60c-src in nerve growth cone membranes. J. Cell Biol. 1990, 111, 1959–1970.
- Park, I.Y.; Powell, R.T.; Tripathi, D.N.; Dere, R.; Ho, T.H.; Blasius, T.L.; Chiang, Y.-C.; Davis, I.J.; Fahey, C.C.; Hacker, K.E.; et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 2016, 166, 950–962.
- Chin, H.G.; Esteve, P.-O.; Ruse, C.; Lee, J.; Schaus, S.E.; Pradhan, S.; Hansen, U. The microtubule-associated histone methyltransferase SET8, facilitated by transcription factor LSF, methylates α-tubulin. J. Biol. Chem. 2020, 295, 4748–4759.
- Ozols, J.; Caron, J.M. Posttranslational modification of tubulin by palmitoylation: II. Identification of sites of palmitoylation. Mol. Biol. Cell 1997, 8, 637–645.
- Song, Y.; Kirkpatrick, L.L.; Schilling, A.B.; Helseth, D.L.; Chabot, N.; Keillor, J.W.; Johnson, G.V.; Brady, S.T. Transglutaminase and Polyamination of Tubulin: Posttranslational Modification for Stabilizing Axonal Microtubules. Neuron 2013, 78, 109–123.
- Hallak, M.E.; Rodriguez, J.; Barra, H.; Caputto, R. Release of tyrosine from tyrosinated tubulin. Some common factors that affect this process and the assembly of tubulin. FEBS Lett. 1977, 73, 147–150.
- Barra, H.S.; Arce, C.A.; Rodriguez, J.A.; Caputto, R. Incorporation of phenylalanine as a single unit into rat brain protein: Reciprocal inhibition by phenylalanine and tyrosine of their respective incorporations. J. Neurochem. 1973, 21, 1241–1251.
- Arce, C.A.; Rodriguez, J.A.; Barra, H.S.; Caputto, R. Incorporation of l-Tyrosine, l-Phenylalanine and l-3,4-Dihydroxyphenylalanine as Single Units into Rat Brain Tubulin. JBIC J. Biol. Inorg. Chem. 1975, 59, 145–149.
- Redeker, V.; Levilliers, N.; Schmitter, J.-M.; Le Caer, J.-P.; Rossier, J.; Adoutte, A.; Bré, M.-H. Polyglycylation of Tubulin: A Posttranslational Modification in Axonemal Microtubules. Science 1994, 266, 1688–1691.
- Bre, M.; Redeker, V.; Quibell, M.; Darmanaden-Delorme, J.; Bressac, C.; Cosson, J.; Huitorel, P.; Schmitter, J.; Rossler, J.; Johnson, T.; et al. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: Widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J. Cell Sci. 1996, 109, 727–738.
- Eddé, B.; Rossier, J.; Le Caer, J.-P.; Desbruyères, E.; Gros, F.; Denoulet, P. Posttranslational Glutamylation of α-tubulin. Science 1990, 247, 83–85.
- E Alexander, J.; Hunt, D.F.; Lee, M.K.; Shabanowitz, J.; Michel, H.; Berlin, S.C.; MacDonald, T.L.; Sundberg, R.J.; I Rebhun, L.; Frankfurter, A. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc. Natl. Acad. Sci. USA 1991, 88, 4685–4689.
- Rüdiger, M.; Plessman, U.; Klöppel, K.-D.; Wehland, J.; Weber, K. Class II tubulin, the major brain β tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 1992, 308, 101–105.
- Ren, Y.; Zhao, J.; Feng, J. Parkin Binds to α/β Tubulin and Increases their Ubiquitination and Degradation. J. Neurosci. 2003, 23, 3316–3324.
- Wang, Q.; Peng, Z.; Long, H.; Deng, X.; Huang, K. Poly-ubiquitylation of α-tubulin at K304 is required for flagellar disassembly in Chlamydomonas. J. Cell Sci. 2019, 132, jcs.229047.
- Rosas-Acosta, G.; Russell, W.K.; Deyrieux, A.; Russell, D.H.; Wilson, V.G. A Universal Strategy for Proteomic Studies of SUMO and Other Ubiquitin-like Modifiers. Mol. Cell. Proteom. 2005, 4, 56–72.
- Paturle, L.; Wehland, J.; Margolis, R.L.; Job, D. Complete separation of tyrosinated, detyrosinated, and nontyrosinatable brain tubulin subpopulations using affinity chromatography. Biochemistry 1989, 28, 2698–2704.
- Paturle-Lafanechere, L.; Edde, B.; Denoulet, P.; Van Dorsselaer, A.; Mazarguil, H.; Le Caer, J.P.; Wehland, J.; Job, D. Characterization of a major brain tubulin variant which cannot be tyrosinated. Biochemistry 1991, 30, 10523–10528.
- Aillaud, C.; Bosc, C.; Saoudi, Y.; Denarier, E.; Peris, L.; Sago, L.; Taulet, N.; Cieren, A.; Tort, O.; Magiera, M.M.; et al. Evidence for new C-terminally truncated variants of α- and β-tubulins. Mol. Biol. Cell 2016, 27, 640–653.
- Hansen, B.K.; Gupta, R.; Baldus, L.; Lyon, D.; Narita, T.; Lammers, M.; Choudhary, C.; Weinert, B.T. Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nat. Commun. 2019, 10, 1055.
- Liu, N.; Xiong, Y.; Li, S.; Ren, Y.; He, Q.; Gao, S.; Zhou, J.; Shui, W. New HDAC6-mediated deacetylation sites of tubulin in the mouse brain identified by quantitative mass spectrometry. Sci. Rep. 2015, 5, 16869.
- Liu, N.; Xiong, Y.; Ren, Y.; Zhang, L.; He, X.; Wang, X.; Liu, M.; Li, D.; Shui, W.; Zhou, J. Proteomic Profiling and Functional Characterization of Multiple Post-Translational Modifications of Tubulin. J. Proteome Res. 2015, 14, 3292–3304.
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic Analysis of Lysine Acetylation Sites in Rat Tissues Reveals Organ Specificity and Subcellular Patterns. Cell Rep. 2012, 2, 419–431.
- Weinert, B.T.; Wagner, S.A.; Horn, H.; Henriksen, P.; Liu, W.R.; Olsen, J.V.; Jensen, L.J.; Choudhary, C. Proteome-Wide Mapping of the Drosophila Acetylome Demonstrates a High Degree of Conservation of Lysine Acetylation. Sci. Signal. 2011, 4, ra48.
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science 2009, 325, 834–840.
- Piroli, G.G.; Manuel, A.M.; Walla, M.D.; Jepson, M.J.; Brock, J.W.C.; Rajesh, M.P.; Tanis, R.M.; Cotham, W.E.; Frizzell, N. Identification of protein succination as a novel modification of tubulin. Biochem. J. 2014, 462, 231–245.
- Ji, S.; Kang, J.G.; Park, S.Y.; Lee, J.; Oh, Y.J.; Cho, J.W. O-GlcNAcylation of tubulin inhibits its polymerization. Amino Acids 2010, 40, 809–818.
- Verhey, K.J.; Gaertig, J. The Tubulin Code. Cell Cycle 2007, 6, 2152–2160.
- Bär, J.; Popp, Y.; Bucher, M.; Mikhaylova, M. Direct and indirect effects of tubulin post-translational modifications on microtubule stability: Insights and regulations. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2022, 15, 119241.
- Kumar, N.; Flavin, M. Preferential action of a brain detyrosinolating carboxypeptidase on polymerized tubulin. J. Biol. Chem. 1981, 256, 7678–7686.
- Shida, T.; Cueva, J.G.; Xu, Z.; Goodman, M.B.; Nachury, M.V. The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. USA 2010, 107, 21517–21522.
- Regnard, C.; Audebert, S.; Desbruyères, É.; Denoulet, P.; Eddé, B. Tubulin Polyglutamylase: Partial Purification and Enzymatic Properties. Biochemistry 1998, 37, 8395–8404.
- Audebert, S.; Desbruyères, E. Reversible Polyglutamylation of Alpha- and Beta-Tubulin and Microtubule Dynamics in Mouse Brain Neurons. Mol. Biol. Cell 1993, 4, 615–626.
- Zhang, Y.; Li, N.; Caron, C.; Matthias, G.; Hess, D.; Khochbin, S.; Matthias, P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003, 22, 1168–1179.
- Szyk, A.; Deaconescu, A.M.; Piszczek, G.; Roll-Mecak, A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 2011, 18, 1250–1258.
- Prota, A.E.; Magiera, M.M.; Kuijpers, M.; Bargsten, K.; Frey, D.; Wieser, M.; Jaussi, R.; Hoogenraad, C.C.; Kammerer, R.A.; Janke, C.; et al. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J. Cell Biol. 2013, 200, 259–270.
- He, K.; Ling, K.; Hu, J. The emerging role of tubulin posttranslational modifications in cilia and ciliopathies. Biophys. Rep. 2020, 6, 1–16.
- Gadadhar, S.; Dadi, H.; Bodakuntla, S.; Schnitzler, A.; Bièche, I.; Rusconi, F.; Janke, C. Tubulin glycylation controls primary cilia length. J. Cell Biol. 2017, 216, 2701–2713.
- Rogowski, K.; Juge, F.; van Dijk, J.; Wloga, D.; Strub, J.-M.; Levilliers, N.; Thomas, D.; Bré, M.-H.; Van Dorsselaer, A.; Gaertig, J.; et al. Evolutionary Divergence of Enzymatic Mechanisms for Posttranslational Polyglycylation. Cell 2009, 137, 1076–1087.
- Wloga, D.; Webster, D.M.; Rogowski, K.; Bré, M.-H.; Levilliers, N.; Jerka-Dziadosz, M.; Janke, C.; Dougan, S.T.; Gaertig, J. TTLL3 Is a Tubulin Glycine Ligase that Regulates the Assembly of Cilia. Dev. Cell 2009, 16, 867–876.
- Koenning, M.; Wang, X.; Karki, M.; Jangid, R.K.; Kearns, S.; Tripathi, D.N.; Cianfrocco, M.; Verhey, K.J.; Jung, S.Y.; Coarfa, C.; et al. Neuronal SETD2 activity links microtubule methylation to an anxiety-like phenotype in mice. Brain 2021, 144, 2527–2540.
- Xie, X.; Wang, S.; Li, M.; Diao, L.; Pan, X.; Chen, J.; Zou, W.; Zhang, X.; Feng, W.; Bao, L. α-TubK40me3 is required for neuronal polarization and migration by promoting microtubule formation. Nat. Commun. 2021, 12, 4113.
- Xu, Z.; Schaedel, L.; Portran, D.; Aguilar, A.; Gaillard, J.; Marinkovich, M.P.; Théry, M.; Nachury, M.V. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 2017, 356, 328–332.
- Martínez-Hernández, J.; Parato, J.; Sharma, A.; Soleilhac, J.-M.; Qu, X.; Tein, E.; Sproul, A.; Andrieux, A.; Goldberg, Y.; Moutin, M.-J.; et al. Crosstalk between acetylation and the tyrosination/detyrosination cycle of α-tubulin in Alzheimer’s disease. Front. Cell Dev. Biol. 2022, 10, 926914.
- Rogowski, K.; van Dijk, J.; Magiera, M.M.; Bosc, C.; Deloulme, J.-C.; Bosson, A.; Peris, L.; Gold, N.D.; Lacroix, B.; Grau, M.B.; et al. A Family of Protein-Deglutamylating Enzymes Associated with Neurodegeneration. Cell 2010, 143, 564–578.
- Magiera, M.M.; Singh, P.; Gadadhar, S.; Janke, C. Tubulin Posttranslational Modifications and Emerging Links to Human Disease. Cell 2018, 173, 1323–1327.
- Yu, I.; Garnham, C.P.; Roll-Mecak, A. Writing and Reading the Tubulin Code. J. Biol. Chem. 2015, 290, 17163–17172.
- Kubo, T.; Yanagisawa, H.-A.; Yagi, T.; Hirono, M.; Kamiya, R. Tubulin Polyglutamylation Regulates Axonemal Motility by Modulating Activities of Inner-Arm Dyneins. Curr. Biol. 2010, 20, 441–445.
- Suryavanshi, S.; Eddé, B.; Fox, L.A.; Guerrero, S.; Hard, R.; Hennessey, T.; Kabi, A.; Malison, D.; Pennock, D.; Sale, W.S.; et al. Tubulin Glutamylation Regulates Ciliary Motility by Altering Inner Dynein Arm Activity. Curr. Biol. 2010, 20, 435–440.
- Johnson, K. The axonemal microtubules of the Chlamydomonas flagellum differ in tubulin isoform content. J. Cell Sci. 1998, 111, 313–320.
