Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Tetraspanin CD81 plays major roles in cell-cell interactions and the regulation of cellular trafficking. This cholesterol-embarking transmembrane protein is a co-receptor for several viruses, including HCV, HIV-1 and Chikungunya virus, which exploits the large extracellular loop (EC2) for cell entry. CD81 is also an anticancer target implicated in cancer cell proliferation and mobility, and in tumor metastasis.
- CD81
- cancer
- drug targeting
- natural products
1. CD81 Structure and Dynamic Architecture
Tetraspanins generally comprise four transmembrane domains and intracellular N and C termini. CD81 is a classical tetraspanin, with a standard transmembrane portion flanked by intra- and extra-cellular domains (Figure 1). The protein functions as a membrane receptor. It can be overexpressed in cultured cells and purified by immune-affinity [1]. At the structural level, the four transmembrane segments (TM1-TM4) of CD81 define a sort of funnel in which a molecule of cholesterol can bind. The funnel is represented by a four-stranded left-handed coiled coil. A large extracellular loop (EC2) caps the funnel, as a lid at the outer membrane leaflet (Figure 1). This large loop EC2 (or LEL) interacts with the vicinal smaller loop EC1 (or SEL) to modulate the conformation of the protein [2]. EC1 contains a small hydrophobic β-strand that packs in a conserved hydrophobic groove of the EC2, which itself is composed of two subdomains, including the δ loop [3][4]. The high-resolution crystal structure of CD81 has confirmed the presence of the two-disulfide bridge in the EC2 domain and a highly-conserved Cys-Cys-Gly motif. The purified full-length human protein has shown a monomeric form, whereas the crystal structure of the extracellular EC2 domain of human CD81 (88 of 236 residues) showed a dimeric assembly [5]. The dimeric form may serve to guide clustering of the different tetraspanin-binding proteins on the cell surface [6]. The organization of CD81 into domains is highly conserved among tetraspanins, notably the transmembrane domain structure, thus facilitating the realization of intra- and inter-molecular interactions and the assembly of the network into the so-called ‘tetraspanin web’ [4][7][8].

Figure 1. CD81 domain organization and structure. (a) Schematic representation of CD81 architecture, with four transmembrane (TM) segments embedded in the membrane bilayer and sequestering a molecule of cholesterol, and two extracellular (EC) loops. The small loop EC1 (or SEL) and the large loop EC2 (or SEL). The disulfur bridges between cysteine residue in EC2 are represented, as well as a palmitoylated cysteine residue contributing the anchoring of the protein into the membrane bilayer. (b) A model of CD81, with EC2 in a closed conformation (based in PDB: 5TCX).
A specific feature of CD81 is its capacity to bind cholesterol. There is a well-defined cholesterol-binding pocket which is important to tune the conformation of the protein and exploited by certain viruses to enter into cells. CD81 serves as co-receptor for many viruses, including the hepatitis C virus (HCV), human immunodeficiency virus type 1 (HIV-1), herpes simplex virus 1 (HSV-1), influenza A virus (IAV), Chikungunya virus and a few others [9][10][11][12][13], and occasionally for certain bacteria, such as Listeria monocytogenes [14][15], and tropical parasites, such as Plasmodium yoelii [16] (Figure 2). The role of CD81 in HCV infection has been largely investigated. A potential allosteric mechanism by which cholesterol binding regulates the conformation of CD81 has been identified recently. The cholesterol-free open form of CD81 exhibits a reduced HCV receptor activity compared to the cholesterol-bound closed conformation which presents an enhanced activity for HCV entry. A conformational switch between the two forms operates, delimiting thus a CD81 cholesterol-sensing mechanism [17]. In its closed conformation, the CD81 extracellular loop EC2 disengages from EC1 and changes conformation. This process prevents the binding of CD81 with its main partner CD19 [18].

Figure 2. Representation of the clustering of tetraspanins to form microdomains at the plasma membrane, associating divers tetraspanins and partner proteins. The tetraspanin web is a dynamic structural organization at the cell surface, including promiscuous and specific interactions [8].
CD81 is a highly dynamic protein, subject to conformational changes which affect its receptor function and its activity in cells. The tetraspanin protein associates with different protein partners and with cholesterol in the membrane, so as to form protein clusters in membrane microdomains, designated tetraspanin webs or tetraspanin-enriched microdomains (Figure 2). These domains correspond to membrane area whereby tetraspanins organize functional higher-order protein complexes, upon interacting with each other and with other transmembrane proteins. CD81 can interact with itself and with other tetraspanins such as CD37, CD53 and CD82 to form individual clusters on the plasma membrane [19]. The sequestering of cholesterol molecules by CD81 within large molecular platforms of proteins induces local conformational changes that perturb the deformability of the membrane [20]. In addition, CD81 is subject to post-transcriptional modifications, notably to a palmitoylation within the intracellular N-terminal segment that is necessary to anchor the protein into lipid rafts [21]. The palmitoylation of cysteine residues of CD81 contributes to the association and anchorage of the protein into the microdomains [4][22]. In contrast, a protein ubiquitination contributes to the removal of the protein from the membrane, through clathrin-mediated endocytosis prior to lysosomal degradation of the protein [23].
2. CD81 Biology, Trafficking and Signaling
Tetraspanins have multiple functions in cells. At the plasma membrane, they promote interactions with other membrane and intracellular proteins and lipids, to shape the organization of membrane domains. They are considered as “molecular facilitators” connecting extracellular and cytoplasmic signaling elements [24]. In this context, CD81 is known to facilitate cells adhesion or fusion in the frame of viral infection [25][26]. The large extracellular loop EC2 (LEL) of CD81 can interact with a variety of proteins, so as to facilitate cellular interaction and capture. Researchers have identified 18 proteins which can interact directly with CD81 (Table 1). The tetraspanin certainly interacts with many other proteins, but true CD81-protein interactions have been evidenced experimentally in a limited number of cases. In other situations, interactions have been suggested based on a colocalization analysis using microscopy for example, but without definitive evidence of direct interaction between CD81 and its colocalization partner. Hereafter, researchers will limit the analysis to the various proteins for which a direct interaction with CD81 has been established (Figure 3).
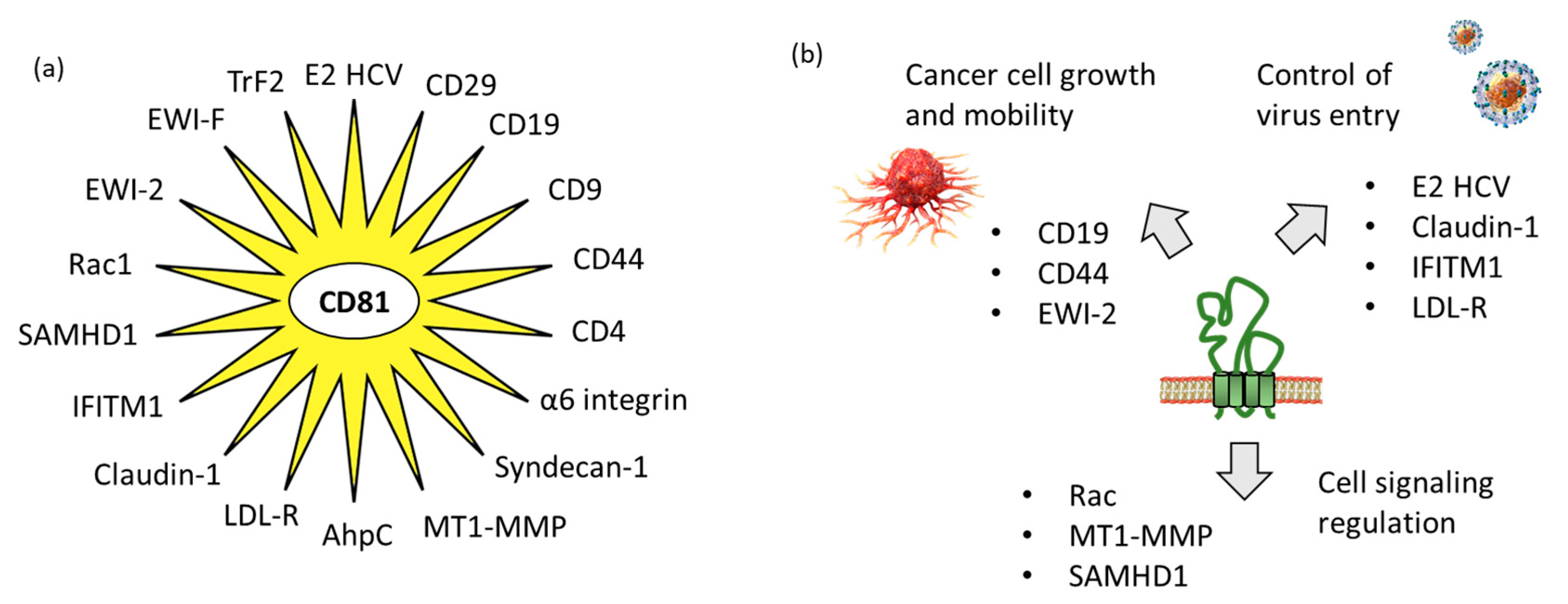
Figure 3. CD81 protein partners. (a) Multiple CD81-interacting proteins have been identified, including ten defined with the support of STRING database “https://cn.string-db.org/ (accessed on 1 March 2023)” and other additional proteins discovered through researchers' analysis of the scientific literature. (b) The partners include proteins interacting with the extracellular loops of CD81, such as proteins implicated in virus entry into cells and proteins involved in cancer cell growth and mobility, but also proteins interacting with the intracellular portions of CD81 for cell signaling.
Table 1. Proteins interacting with CD81.
| Proteins | Types | Interaction and Effects | References |
|---|---|---|---|
| E2 HCV | Viral protein | The transmembrane E2 glycoprotein of HCV utilizes CD81 as a coreceptor for cell entry. | [27][28][29] |
| Claudin-1 | Tight junction protein | The interaction CD81-Claudin1 contributes to HCV infection. | [30] |
| IFITM1 | Tight junction protein | IFITM1 interacts with CD81 to limit HCV entry. | [31][32] |
| LDL-R | Membrane receptor |
Interplay between CD81, LDL-R and PCSK9, to control HCV entry into hepatic cells. | [33][34] |
| CD19 | Membrane receptor |
CD81 and CD19 are core subunits of the B cell co-receptor complex. CD81 controls CD19 export activity, via a dynamically regulated process upon B cell activation. | [35] |
| EWI-2 (IgSF8, PGRL, CD316) |
Signaling protein |
The CD81/EWI-2 interaction contributes to the tetraspanin web and plays role in cancer cell growth and motility, and in HCV entry. | [36][37][38] |
| EWI-F (FPRP, CD9P-1) |
Signaling protein |
Complexes formed between EWI-F and CD81 (and CD9) play a role in the fusion of myotubes, which are essential elements of muscle architecture. | [39] |
| CD44 | Adhesion molecule |
The interaction between CD81 and CD44, through their extracellular regions, promotes tumor cell cluster formation and lung metastasis of triple negative breast cancer. | [40] |
| α6 integrin | Adhesion molecule |
In male germ cells, CD81 interacts with α6 integrin subunit (which forms a dimer with β4 integrin). The complex plays a role in sperm maturation. | [41] |
| β1 integrin (CD29) |
Adhesion molecule |
Radiation was found to induce CD29/CD81 complex formation, thereby increasing the cellular uptake of exosomes. | [42] |
| Rac1 | Small GTPase | Interaction of Rac with the C-terminal cytoplasmic portion of CD81 to regulate cell motility. Also has a role in bacterial infection. | [15] |
| CD4 | Cell surface antigen |
CD81 interacts with CD4 dimers concentrated in tetraspanin-enriched microdomains. | [43] |
| CD9 | Tetraspanin | Tetraspanins CD9 and CD81 are involved in tetraspanin web formation in sperm. Molecular modelling suggests protein-protein interactions during sperm-egg membrane fusion. | [44] |
| SAMHD1 | Enzyme | CD81 interacts with the deoxynucleoside triphosphate phosphohydrolase SAMHD1 and regulates its expression. The interaction promotes the proteasome-dependent degradation of SAMHD1. It is one of the metabolic regulators of HIV-1 replication. | [45] |
| MT1-MMP | Enzyme | Several tetraspanins, including CD81, associate with the membrane-type 1 matrix metalloproteinase (MT1-MMP) to regulate its cell surface localization and its function (notably its capacity to activate pro-MMP-2). | [46] |
| Syndecan-1 | Proteoglycan | Knockdown of Syndecan-1 and CD81 inhibits HCV infection, suggesting their cooperative action. A direct interaction between the two proteins has been evidenced (using a proximity ligation assay). | [47] |
| AhpC | Enzyme | The mycobacterial enzyme alkyl hydroperoxide reductase C (AhpC) interacts with CD81-LEL to promote uptake of the pathogen by host cells. | [48] |
| TfR2 | Membrane receptor |
Transferrin receptor 2 (TfR2) is a binding partner for CD81. The interaction triggers RfR2 degradation by the ubiquitin E3 ligase GRAIL. | [49] |
2.1. CD81 Protein Partners Implicated in Virus Uptake
One of the prominent binders of CD81-LEL is the E2 envelope glycoprotein which plays a key role in the attachment and entry of the HCV virus into infected cells. CD81 increases the interaction of E2 with membranes and triggers a conformational change in E2 necessary for subsequent membrane fusion [27]. In fact, via E2 the virus utilizes different proteins as co-receptors, in particular, the tight-junction proteins claudin-1 and occludin together with CD81 and the protein SR-B1 (scavenger receptor class B member 1, encoded by gene SCARB1) [50]. SR-B1 is an 82-kDa transmembrane glycoprotein playing an important role in the regulation of cholesterol exchange between cells and high-density lipoproteins. Does CD81 directly interact with SR-B1? There is some evidence for that, notably data indicating that HCV E2 links a soluble form of CD81 and SR-B1 protein together. This physical neighboring could explain why both CD81 and SR-B1 are indispensable factors for HCV infection [51]. Moreover, the physical interaction between CD81 and claudin-1 has been firmly demonstrated. Claudin-1 oligomers associate with CD81 to form complexes playing a role in HCV infection [30][52]. In the same vein, a physical interaction has been evidenced between CD81 and protein IFITM1 (interferon-induced transmembrane 1) which is a hepatocyte tight junction protein implicated in HCV entry. The interaction of IFITM1 with HCV coreceptors CD81 and occludin disrupts the process of viral entry [31][32]. The low-density lipoprotein receptor (LDL-R) is involved also in viral entry. The formation of complexes between CD81, LDL-R and the serine proteinase PCSK9 (proprotein convertase subtilisin kexin type 9) has been observed [33]. PCSK9 enhances the degradation of the LDL-R and modulates liver CD81 levels [34]. However, the targeting of PCSK9 (implicated in cholesterol metabolism) with a mAb (alirocumab) does not affect CD81 and does not modulate HCV entry in hepatic cells [53].
2.2. CD81 Protein Partners Implicated in Tumor Growth and Dissemination
Another essential partner of CD81 is the B-lymphocyte coreceptor CD19, which is a key activator of the PI3K pathway and a prominent tumor-associated antigen. CD19 is a B/plasma cell-lineage marker and an essential target to combat B-cell malignancies, through the design of anti-CD19 chimeric antigen receptor (CAR) T-cell therapies [54]. For example, the CAR T-cell therapy axicabtagene ciloleucel has been recently approved for the treatment of relapsed or refractory follicular lymphoma [55]. Binding of CD19 to CD81 induces opening of the ectodomain and a reorganization of transmembrane helices, resulting in the occlusion of the cholesterol binding pocket [35]. This CD81-CD19 interaction, dynamically regulated upon B cell activation [56], is essential to the correct exposure of CD19 on the surface of B cells. Loss of CD81 expression results in an intracellular accumulation of CD19 [57]. Similarly, a mutation in the CD81 gene leads to the expression of a truncated protein which does not enable CD19 maturation and cell surface expression [58]. The CD19-complex consisting of CD19, CD81, CD21 (also known as CR2, for complement receptor 2) and CD225 acts as a co-receptor to the B cell receptor (BCR). This complex cannot form properly when CD81 is mutated, causing severe diseases, but fortunately CD81 mutations are extremely rare in humans [59]. CD81 is an essential T cell costimulatory molecule. Its co-stimulation enhances naive T cell activation and largely modulates the activation of chimeric antigen receptors (CAR) [60].
CD81 associates with the two EWI proteins EWI-2 and EWI-F which are also partners of tetraspanin CD9 (Table 1). These two interactions are important because they have implications for cancer. EWI-2 (also called PGRL in mouse (prostaglandin regulatory-like protein)) is a regulator of both CD81 and CD9 functions [61], acting as a sequester to prevent the two tetraspanins from providing support to the TGF-β signaling. When EWI-2 binds CD9/CD81, the tetraspanins can no longer support the association of TGF-β receptors 1 and 2 (TβR2-TβR1). This signaling pathway is largely involved in melanoma growth, invasion and metastasis [36][62]. The suppression of CD81 with a shRNA in mesenchymal breast cancer has been shown to reduce primary tumor growth, extravasation and lung metastasis in vivo [63]. Therefore, the pharmacological blockade of CD81 could be an option to alter the malignant process.
The CD81/EWI-2 interaction has multiple functions, notably acting as a linker of the tetraspanin web to the actin cytoskeleton [64], and this interaction plays roles in different pathologies. CD81/EWI-2 interaction is a regulatory factor for glioblastoma cell growth and motility [37], but also for HCV and HIV infection [38][65]. Notably, the HIV-1 virus uses CD81-lined vesicle structures to infect astrocytes, and then these energy-consuming glial cells support trans-infection of HIV-1 to T-cells [66]. The virus uses CD81 as a rheostat to control different stages of the infection via interaction with different proteins such as EWI-2, but also the deoxynucleoside triphosphate phosphohydrolase SAMHD1 (sterile alpha motif and histidine aspartic acid domain containing protein 1) [45]. Through direct binding to SAMHD1, CD81 regulates the expression of the protein by promoting its proteasome-dependent degradation. This mechanism is implicated in the control of HIV-1 replication [45]. The viral restriction factor SAMHD1 is also considered as an anticancer target. The protein is frequently upregulated in cytarabine (Ara-C)-resistant AML [67]. SAMHD1 inhibitors are being developed for the sensitization of leukemia cells to nucleoside analogue-based therapy [68]. The link between SAMHD1 and CD81 may explain, at least in part, why CD81 is an adverse prognostic marker in AML [69].
The interaction of tetraspanins with the cell surface protein EWI-F (also known as CD9P-1 (CD9 Partner-1) or FPRP (F2alpha prostaglandin receptor regulatory protein)) has been well characterized. EWI-F is an immunoglobulin domain molecule which chiefly interacts with CD9 to form a 2:2 tetrameric arrangement implicated in the formation of tetraspanin-enriched microdomains [70]. In myoblasts, both CD9 or CD81 associate with EWI-F and the complex plays a role in muscle architecture (fusion of myotubes) and muscle regeneration [39]. Here again, the CD9P-1/tetraspanin complex functions as a regulator of cell motility [71]. Interestingly, a truncated form of EWI-F/CD9P-1, designated GS-168AT2, produced in human endothelial cells has been shown to inhibit angiogenesis and cell migration. GS-168AT2 corresponds to the sequence by which CD9P-1 physiologically associates with CD81 [72]. GS-168AT2 binds CD9 and CD81 and displays antitumor activity in vivo, associated with a downregulation of CD9 but not of CD81 [73]. This work suggested that the pharmacological modulation of the tetraspanin web could represent a new anti-angiogenic strategy [74]. The interaction between CD81 and CD9P-1 could also be exploited in the field of parasitic pathologies, because it has been shown that binding to and regulation of CD81 function by CD9P-1 represents a negative regulator of infection by Plasmodium yoelii (a malaria pathogen in rodent species) [16]. CD81 is implicated in the uptake of different pathogenic agents. A recent study highlighted the key role of CD81 in the uptake of pathogenic mycobacteria Mycobacterium abscessus through interaction with alkyl hydroperoxide reductase C (AhpC), a peroxiredoxin enzyme able to decompose several kinds of hydroperoxides [48]. The targeting of CD81 could be further exploited to combat different infectious diseases.
The hyaluronan-binding protein CD44 (a hyaladherin) is also a binding partner for CD81. A complex formation between these two membrane proteins has been evidenced recently [40]. They interact with each other through their extracellular regions and this recognition facilitates the formation of a tumor cell cluster and lung metastasis of triple negative breast cancer (TNBC). This interaction confirms the key role of CD81 in the migration and invasion of TNBC cells previously suggested by a proteomic analysis [75]. The invasion of TNBC cell lines can be halted with the use of a specific anti-human CD81 antibody (5A6) [76].
The CD81 interactome includes also the small GTPase Rac and the interaction plays roles in cancer cell motility and exosome formation [77]. The interaction of Rac (Rac1) with the C-terminal cytoplasmic domain of CD81 regulates the GTPase activity [78][79] and the modulation of Rac1 GTPase functions represents a key anticancer mechanism [80]. This tetraspanin-dependent regulation of cell motility raises an opportunity to control cancer cell migration with C81-targeted drugs. In addition, the CD81-Rac interaction plays a regulatory role in the innate and adaptive immunity against bacterial infection [15]. The signaling activity could be blocked with small molecules targeting the intracellular part of CD81 but probably also via targeting the extracellular protein loop.
Finally, researchers can evoke the association of tetraspanins with various integrins to modulate their function. Both tetraspanins CD9 and CD81 are a potential partner of αV integrin, at least in the testicular tissue where they are implicated in sperm development [81][82][83]. These two tetraspanins were also shown to link the cell adhesion molecule JAM-A to αvβ5 integrin and thus to play a role in the regulation of cell motility [84]. CD81 forms a complex with αV/β1 and αV/β5 integrins [85]. Integrins bind to tetraspanins such as CD81 via interaction with the constant region of the EC2 domain [84]. There are other integrins capable of interacting with CD81, such as α3β1 integrin, CD29, and others [42][85][86].
Altogether, researcher' biological network analysis points out 17 protein partners for CD81 (Table 1). Most of them interact via the extracellular domain- of the tetraspanin, but in some cases the interaction concerns the intracellular portion, as depicted in Figure 3. There are probably many other partners, associated with CD81 itself or with CD81-containing tetraspanin platforms. A proteomic analysis mapped 33 host protein interactions of CD81 in primary human liver and hepatoma cells, including for example protein CAPN5 (calpain-5) and ubiquitin ligase CBLB (Casitas B-lineage lymphoma proto-oncogene B), capable of forming a complex with CD81 and implicated in HCV entry [87]. CD81 presents a dynamic expression profile and the multiplicity of partners is not surprising for a protein implicated in intercellular communication. Therefore, therapeutic molecules interacting with CD81 could serve as interrupters or regulators of different pathways and cellular processes. The next section provides an overview of CD81-binding molecules, large and small.
This entry is adapted from the peer-reviewed paper 10.3390/cancers15072186
References
- Takayama, H.; Chelikani, P.; Reeves, P.J.; Zhang, S.; Khorana, H.G. High-level expression, single-step immunoaffinity purification and characterization of human tetraspanin membrane protein CD81. PLoS ONE 2008, 3, e2314.
- Zimmerman, B.; Kelly, B.; McMillan, B.J.; Seegar, T.C.M.; Dror, R.O.; Kruse, A.C.; Blacklow, S.C. Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket. Cell 2016, 167, 1041–1051.
- Seigneuret, M.; Delaguillaumie, A.; Lagaudrière-Gesbert, C.; Conjeaud, H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 2001, 276, 40055–40064.
- Seigneuret, M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: Conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 2006, 90, 212–227.
- Kitadokoro, K.; Ponassi, M.; Galli, G.; Petracca, R.; Falugi, F.; Grandi, G.; Bolognesi, M. Subunit association and conformational flexibility in the head subdomain of human CD81 large extracellular loop. Biol. Chem. 2002, 383, 1447–1452.
- Kitadokoro, K.; Bordo, D.; Galli, G.; Petracca, R.; Falugi, F.; Abrignani, S.; Grandi, G.; Bolognesi, M. CD81 extracellular domain 3D structure: Insight into the tetraspanin superfamily structural motifs. EMBO J. 2001, 20, 12–18.
- Stipp, C.S.; Kolesnikova, T.V.; Hemler, M.E. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 2003, 28, 106–112.
- van Deventer, S.J.; Dunlock, V.E.; van Spriel, A.B. Molecular interactions shaping the tetraspanin web. Biochem. Soc. Trans. 2017, 45, 741–750.
- New, C.; Lee, Z.Y.; Tan, K.S.; Wong, A.H.; Wang, Y.; Tran, T. Tetraspanins: Host Factors in Viral Infections. Int. J. Mol. Sci. 2021, 22, 11609.
- Florin, L.; Lang, T. Tetraspanin Assemblies in Virus Infection. Front. Immunol. 2018, 9, 1140.
- Qiao, Y.; Yan, Y.; Tan, K.S.; Tan, S.S.L.; Seet, J.E.; Arumugam, T.V.; Chow, V.T.K.; Wang, Y.; Tran, T. CD151, a novel host factor of nuclear export signaling in influenza virus infection. J. Allergy Clin. Immunol. 2018, 141, 1799–1817.
- Benayas, B.; Sastre, I.; López-Martín, S.; Oo, A.; Kim, B.; Bullido, M.J.; Aldudo, J.; Yáñez-Mó, M. Tetraspanin CD81 regulates HSV-1 infection. Med. Microbiol. Immunol. 2020, 209, 489–498.
- Lasswitz, L.; Zapatero-Belinchón, F.J.; Moeller, R.; Hülskötter, K.; Laurent, T.; Carlson, L.A.; Goffinet, C.; Simmons, G.; Baumgärtner, W.; Gerold, G. The Tetraspanin CD81 Is a Host Factor for Chikungunya Virus Replication. mBio. 2022, 13, e0073122.
- Tham, T.N.; Gouin, E.; Rubinstein, E.; Boucheix, C.; Cossart, P.; Pizarro-Cerda, J. Tetraspanin CD81 is required for Listeria monocytogenes invasion. Infect. Immun. 2010, 78, 204–209.
- Martínez del Hoyo, G.; Ramírez-Huesca, M.; Levy, S.; Boucheix, C.; Rubinstein, E.; Minguito de la Escalera, M.; González-Cintado, L.; Ardavín, C.; Veiga, E.; Yáñez-Mó, M.; et al. CD81 controls immunity to Listeria infection through rac-dependent inhibition of proinflammatory mediator release and activation of cytotoxic T cells. J. Immunol. 2015, 194, 6090–6101.
- Charrin, S.; Yalaoui, S.; Bartosch, B.; Cocquerel, L.; Franetich, J.F.; Boucheix, C.; Mazier, D.; Rubinstein, E.; Silvie, O. The Ig domain protein CD9P-1 down-regulates CD81 ability to support Plasmodium yoelii infection. J. Biol. Chem. 2009, 284, 31572–31578.
- Palor, M.; Stejskal, L.; Mandal, P.; Lenman, A.; Alberione, M.P.; Kirui, J.; Moeller, R.; Ebner, S.; Meissner, F.; Gerold, G.; et al. Cholesterol sensing by CD81 is important for hepatitis C virus entry. J. Biol. Chem. 2020, 295, 16931–16948.
- Yang, Y.; Liu, X.R.; Greenberg, Z.J.; Zhou, F.; He, P.; Fan, L.; Liu, S.; Shen, G.; Egawa, T.; Gross, M.L.; et al. Open conformation of tetraspanins shapes interaction partner networks on cell membranes. EMBO J. 2020, 39, e105246.
- Zuidscherwoude, M.; Göttfert, F.; Dunlock, V.M.; Figdor, C.G.; van den Bogaart, G.; van Spriel, A.B. The tetraspanin web revisited by super-resolution microscopy. Sci. Rep. 2015, 5, 12201.
- Caparotta, M.; Masone, D. Cholesterol plays a decisive role in tetraspanin assemblies during bilayer deformations. Biosystems. 2021, 209, 104505.
- Cherukuri, A.; Carter, R.H.; Brooks, S.; Bornmann, W.; Finn, R.; Dowd, C.S.; Pierce, S.K. B cell signaling is regulated by induced palmitoylation of CD81. J. Biol. Chem. 2004, 279, 31973–31982.
- Zhu, Y.Z.; Luo, Y.; Cao, M.M.; Liu, Y.; Liu, X.Q.; Wang, W.; Wu, D.G.; Guan, M.; Xu, Q.Q.; Ren, H.; et al. Significance of palmitoylation of CD81 on its association with tetraspanin-enriched microdomains and mediating hepatitis C virus cell entry. Virology 2012, 429, 112–123.
- Hosokawa, K.; Ishimaru, H.; Watanabe, T.; Fujimuro, M. The Lysosome Pathway Degrades CD81 on the Cell Surface by Poly-ubiquitination and Clathrin-Mediated Endocytosis. Biol. Pharm. Bull. 2020, 43, 540–545.
- Orinska, Z.; Hagemann, P.M.; Halova, I.; Draber, P. Tetraspanins in the regulation of mast cell function. Med. Microbiol. Immunol. 2020, 209, 531–543.
- Lu, C.; Feng, Y.; Sun, X.; Li, N.; Kuang, D.; Wang, W.; Tong, P.; Han, Y.; Xia, X.; Dai, J. Tree shrew bone marrow-derived mesenchymal stem cells express CD81, OCLN, and miR-122, facilitating the entire hepatitis C virus life cycle. J. Med. Virol. 2020; online ahead of print.
- Liao, L.; Wu, Z.; Chen, W.; Zhang, H.; Li, A.; Yan, Y.; Xie, Z.; Li, H.; Lin, W.; Ma, J.; et al. Anti-CD81 antibody blocks vertical transmission of avian leukosis virus subgroup J. Vet. Microbiol. 2022, 264, 109293.
- Kumar, A.; Hossain, R.A.; Yost, S.A.; Bu, W.; Wang, Y.; Dearborn, A.D.; Grakoui, A.; Cohen, J.I.; Marcotrigiano, J. Structural insights into hepatitis C virus receptor binding and entry. Nature 2021, 598, 521–525.
- Ayub, H.; Clare, M.; Milic, I.; Chmel, N.P.; Böning, H.; Devitt, A.; Krey, T.; Bill, R.M.; Rothnie, A.J. CD81 extracted in SMALP nanodiscs comprises two distinct protein populations within a lipid environment enriched with negatively charged headgroups. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183419.
- Ströh, L.J.; Krey, T. HCV Glycoprotein Structure and Implications for B-Cell Vaccine Development. Int. J. Mol. Sci. 2020, 21, 6781.
- Bonander, N.; Jamshad, M.; Oberthür, D.; Clare, M.; Barwell, J.; Hu, K.; Farquhar, M.J.; Stamataki, Z.; Harris, H.J.; Dierks, K.; et al. Production, purification and characterization of recombinant, full-length human claudin-1. PLoS ONE 2013, 8, e64517.
- Wilkins, C.; Woodward, J.; Lau, D.T.; Barnes, A.; Joyce, M.; McFarlane, N.; McKeating, J.A.; Tyrrell, D.L.; Gale, M., Jr. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 2013, 57, 461–469.
- Narayana, S.K.; Helbig, K.J.; McCartney, E.M.; Eyre, N.S.; Bull, R.A.; Eltahla, A.; Lloyd, A.R.; Beard, M.R. The Interferon-induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry. J. Biol. Chem. 2015, 290, 25946–25959.
- Le, Q.T.; Blanchet, M.; Seidah, N.G.; Labonté, P. Plasma Membrane Tetraspanin CD81 Complexes with Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) and Low-Density Lipoprotein Receptor (LDLR), and Its Levels Are Reduced by PCSK9. J. Biol. Chem. 2015, 290, 23385–23400.
- Bridge, S.H.; Sheridan, D.A.; Felmlee, D.J.; Crossey, M.M.; Fenwick, F.I.; Lanyon, C.V.; Dubuc, G.; Seidah, N.G.; Davignon, J.; Thomas, H.C.; et al. PCSK9, apolipoprotein E and lipoviral particles in chronic hepatitis C genotype 3: Evidence for genotype-specific regulation of lipoprotein metabolism. J. Hepatol. 2015, 62, 763–770.
- Susa, K.J.; Rawson, S.; Kruse, A.C.; Blacklow, S.C. Cryo-EM structure of the B cell co-receptor CD19 bound to the tetraspanin CD81. Science 2021, 371, 300–305.
- Wang, H.X.; Sharma, C.; Knoblich, K.; Granter, S.R.; Hemler, M.E. EWI-2 negatively regulates TGF-β signaling leading to altered melanoma growth and metastasis. Cell. Res. 2015, 25, 370–385.
- Kolesnikova, T.V.; Stipp, C.S.; Rao, R.M.; Lane, W.S.; Luscinskas, F.W.; Hemler, M.E. EWI-2 modulates lymphocyte integrin alpha4beta1 functions. Blood 2004, 103, 3013–3019.
- Montpellier, C.; Tews, B.A.; Poitrimole, J.; Rocha-Perugini, V.; D’Arienzo, V.; Potel, J.; Zhang, X.A.; Rubinstein, E.; Dubuisson, J.; Cocquerel, L. Interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection. J. Biol. Chem. 2011, 286, 13954–13965.
- Charrin, S.; Latil, M.; Soave, S.; Polesskaya, A.; Chrétien, F.; Boucheix, C.; Rubinstein, E. Normal muscle regeneration requires tight control of muscle cell fusion by tetraspanins CD9 and CD81. Nat. Commun. 2013, 4, 1674.
- Ramos, E.K.; Tsai, C.F.; Jia, Y.; Cao, Y.; Manu, M.; Taftaf, R.; Hoffmann, A.D.; El-Shennawy, L.; Gritsenko, M.A.; Adorno-Cruz, V.; et al. Machine learning-assisted elucidation of CD81-CD44 interactions in promoting cancer stemness and extracellular vesicle integrity. Elife 2022, 11, e82669.
- Jankovicova, J.; Frolikova, M.; Palenikova, V.; Valaskova, E.; Cerny, J.; Secova, P.; Bartokova, M.; Horovska, L.; Manaskova-Postlerova, P.; Antalikova, J.; et al. Expression and distribution of CD151 as a partner of alpha6 integrin in male germ cells. Sci. Rep. 2020, 10, 4374.
- Hazawa, M.; Tomiyama, K.; Saotome-Nakamura, A.; Obara, C.; Yasuda, T.; Gotoh, T.; Tanaka, I.; Yakumaru, H.; Ishihara, H.; Tajima, K. Radiation increases the cellular uptake of exosomes through CD29/CD81 complex formation. Biochem. Biophys. Res. Commun. 2014, 446, 1165–1171.
- Fournier, M.; Peyrou, M.; Bourgoin, L.; Maeder, C.; Tchou, I.; Foti, M. CD4 dimerization requires two cysteines in the cytoplasmic domain of the molecule and occurs in microdomains distinct from lipid rafts. Mol. Immunol. 2010, 47, 2594–2603.
- Frolikova, M.; Manaskova-Postlerova, P.; Cerny, J.; Jankovicova, J.; Simonik, O.; Pohlova, A.; Secova, P.; Antalikova, J.; Dvorakova-Hortova, K. CD9 and CD81 Interactions and Their Structural Modelling in Sperm Prior to Fertilization. Int. J. Mol. Sci. 2018, 19, 1236.
- Rocha-Perugini, V.; Suárez, H.; Álvarez, S.; López-Martín, S.; Lenzi, G.M.; Vences-Catalán, F.; Levy, S.; Kim, B.; Muñoz-Fernández, M.A.; Sánchez-Madrid, F.; et al. CD81 association with SAMHD1 enhances HIV-1 reverse transcription by increasing dNTP levels. Nat. Microbiol. 2017, 2, 1513–1522.
- Schröder, H.M.; Hoffmann, S.C.; Hecker, M.; Korff, T.; Ludwig, T. The tetraspanin network modulates MT1-MMP cell surface trafficking. Int. J. Biochem. Cell. Biol. 2013, 45, 1133–1144.
- Grigorov, B.; Reungoat, E.; Gentil Dit Maurin, A.; Varbanov, M.; Blaising, J.; Michelet, M.; Manuel, R.; Parent, R.; Bartosch, B.; Zoulim, F.; et al. Hepatitis C virus infection propagates through interactions between Syndecan-1 and CD81 and impacts the hepatocyte glycocalyx. Cell. Microbiol. 2017, 19, e12711.
- Karam, J.; Blanchet, F.P.; Vivès, É.; Boisguérin, P.; Boudehen, Y.M.; Kremer, L.; Daher, W. Mycobacterium abscessus alkyl hydroperoxide reductase C promotes cell invasion by binding to tetraspanin CD81. iScience 2023, 26, 106042.
- Chen, J.; Enns, C.A. CD81 promotes both the degradation of transferrin receptor 2 (TfR2) and the Tfr2-mediated maintenance of hepcidin expression. J. Biol. Chem. 2015, 290, 7841–7850.
- Khan, A.G.; Whidby, J.; Miller, M.T.; Scarborough, H.; Zatorski, A.V.; Cygan, A.; Price, A.A.; Yost, S.A.; Bohannon, C.D.; Jacob, J.; et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 2014, 509, 381–384.
- Heo, T.H.; Lee, S.M.; Bartosch, B.; Cosset, F.L.; Kang, C.Y. Hepatitis C virus E2 links soluble human CD81 and SR-B1 protein. Virus Res. 2006, 121, 58–64.
- Davis, C.; Harris, H.J.; Hu, K.; Drummer, H.E.; McKeating, J.A.; Mullins, J.G.; Balfe, P. In silico directed mutagenesis identifies the CD81/claudin-1 hepatitis C virus receptor interface. Cell. Microbiol. 2012, 14, 1892–1903.
- Ramanathan, A.; Gusarova, V.; Stahl, N.; Gurnett-Bander, A.; Kyratsous, C.A. Alirocumab, a Therapeutic Human Antibody to PCSK9, Does Not Affect CD81 Levels or Hepatitis C Virus Entry and Replication into Hepatocytes. PLoS ONE 2016, 11, e0154498.
- Sermer, D.; Elavalakanar, P.; Abramson, J.S.; Palomba, M.L.; Salles, G.; Arnason, J. Targeting CD19 for diffuse large B cell lymphoma in the era of CARs: Other modes of transportation. Blood Rev. 2023, 57, 101002.
- Cohen, J.A.; Ghobadi, A. Axicabtagene ciloleucel for the treatment of relapsed or refractory follicular lymphoma. Expert Rev. Anticancer Ther. 2022, 22, 903–914.
- Susa, K.J.; Seegar, T.C.; Blacklow, S.C.; Kruse, A.C. A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking. Elife 2020, 9, e52337.
- Velasquez, M.P.; Gottschalk, S. Targeting CD19: The good, the bad, and CD81. Blood 2017, 129, 9–10.
- Vences-Catalán, F.; Rajapaksa, R.; Srivastava, M.K.; Marabelle, A.; Kuo, C.C.; Levy, R.; Levy, S. Tetraspanin CD81 promotes tumor growth and metastasis by modulating the functions of T regulatory and myeloid-derived suppressor cells. Cancer Res. 2015, 75, 4517–4526.
- Yang, L.; Liu, P.; Du, H.; Chen, R.; Zhou, B.; Li, Y.; Zhou, L.; Wang, X.; Liu, C.; Ding, Y.; et al. Novel CD81 Mutations in a Chinese Patient Led to IgA Nephropathy and Impaired BCR Signaling. J. Clin. Immunol. 2022, 42, 1672–1684.
- Schultz, L.M.; Czerwinski, D.K.; Levy, R.; Levy, S. CD81 costimulation skews CAR transduction toward naive T cells. Proc. Natl. Acad. Sci. USA 2022, 119, e1910844119.
- Stipp, C.S.; Kolesnikova, T.V.; Hemler, M.E. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 2001, 276, 40545–40554.
- Wang, H.X.; Hemler, M.E. Novel impact of EWI-2, CD9, and CD81 on TGF-β signaling in melanoma. Mol. Cell. Oncol. 2015, 2, e1030536.
- Uretmen Kagiali, Z.C.; Sanal, E.; Karayel, Ö.; Polat, A.N.; Saatci, Ö.; Ersan, P.G.; Trappe, K.; Renard, B.Y.; Önder, T.T.; Tuncbag, N.; et al. Systems-level Analysis Reveals Multiple Modulators of Epithelial-mesenchymal Transition and Identifies DNAJB4 and CD81 as Novel Metastasis Inducers in Breast Cancer. Mol. Cell. Proteomics. 2019, 18, 1756–1771.
- Sala-Valdés, M.; Ursa, A.; Charrin, S.; Rubinstein, E.; Hemler, M.E.; Sánchez-Madrid, F.; Yáñez-Mó, M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J. Biol. Chem. 2006, 281, 19665–19675.
- Whitaker, E.E.; Matheson, N.J.; Perlee, S.; Munson, P.B.; Symeonides, M.; Thali, M. EWI-2 Inhibits Cell-Cell Fusion at the HIV-1 Virological Presynapse. Viruses. 2019, 11, 1082.
- Gray, L.R.; Turville, S.G.; Hitchen, T.L.; Cheng, W.J.; Ellett, A.M.; Salimi, H.; Roche, M.J.; Wesselingh, S.L.; Gorry, P.R.; Churchill, M.J. HIV-1 entry and trans-infection of astrocytes involves CD81 vesicles. PLoS ONE 2014, 9, e90620.
- Zhang, F.; Sun, J.; Tang, X.; Liang, Y.; Jiao, Q.; Yu, B.; Dai, Z.; Yuan, X.; Li, J.; Yan, J.; et al. Stabilization of SAMHD1 by NONO is crucial for Ara-C resistance in AML. Cell. Death Dis. 2022, 13, 590.
- Rothenburger, T.; Thomas, D.; Schreiber, Y.; Wratil, P.R.; Pflantz, T.; Knecht, K.; Digianantonio, K.; Temple, J.; Schneider, C.; Baldauf, H.M.; et al. Differences between intrinsic and acquired nucleoside analogue resistance in acute myeloid leukaemia cells. J. Exp. Clin. Cancer Res. 2021, 40, 317.
- Boyer, T.; Guihard, S.; Roumier, C.; Peyrouze, P.; Gonzales, F.; Berthon, C.; Quesnel, B.; Preudhomme, C.; Behal, H.; Duhamel, A.; et al. Tetraspanin CD81 is an adverse prognostic marker in acute myeloid leukemia. Oncotarget 2016, 7, 62377–62385.
- Oosterheert, W.; Xenaki, K.T.; Neviani, V.; Pos, W.; Doulkeridou, S.; Manshande, J.; Pearce, N.M.; Kroon-Batenburg, L.M.; Lutz, M.; van Bergen En Henegouwen, P.M.; et al. Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F. Life Sci. Alliance. 2020, 3, e202000883.
- Chambrion, C.; Le Naour, F. The tetraspanins CD9 and CD81 regulate CD9P1-induced effects on cell migration. PLoS ONE 2010, 5, e11219.
- Colin, S.; Guilmain, W.; Creoff, E.; Schneider, C.; Steverlynck, C.; Bongaerts, M.; Legrand, E.; Vannier, J.P.; Muraine, M.; Vasse, M.; et al. A truncated form of CD9-partner 1 (CD9P-1), GS-168AT2, potently inhibits in vivo tumour-induced angiogenesis and tumour growth. Br. J. Cancer. 2011, 105, 1002–1011.
- Guilmain, W.; Colin, S.; Legrand, E.; Vannier, J.P.; Steverlynck, C.; Bongaerts, M.; Vasse, M.; Al-Mahmood, S. CD9P-1 expression correlates with the metastatic status of lung cancer, and a truncated form of CD9P-1, GS-168AT2, inhibits in vivo tumour growth. Br. J. Cancer. 2011, 104, 496–504.
- Vasse, M.; Colin, S.; Guilmain, W.; Creoff, E.; Muraine, M.; Vannier, J.P.; Al-Mahmood, S. . Ann. Pharm. Fr. 2015, 73, 100–107.
- Li, S.; Li, X.; Yang, S.; Pi, H.; Li, Z.; Yao, P.; Zhang, Q.; Wang, Q.; Shen, P.; Li, X.; et al. Proteomic Landscape of Exosomes Reveals the Functional Contributions of CD151 in Triple-Negative Breast Cancer. Mol. Cell. Proteomics. 2021, 20, 100121.
- Vences-Catalán, F.; Rajapaksa, R.; Kuo, C.C.; Miller, C.L.; Lee, A.; Ramani, V.C.; Jeffrey, S.S.; Levy, R.; Levy, S. Targeting the tetraspanin CD81 reduces cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2021, 118, e2018961118.
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661.
- Quast, T.; Eppler, F.; Semmling, V.; Schild, C.; Homsi, Y.; Levy, S.; Lang, T.; Kurts, C.; Kolanus, W. CD81 is essential for the formation of membrane protrusions and regulates Rac1-activation in adhesion-dependent immune cell migration. Blood 2011, 118, 1818–1827.
- Tejera, E.; Rocha-Perugini, V.; López-Martín, S.; Pérez-Hernández, D.; Bachir, A.I.; Horwitz, A.R.; Vázquez, J.; Sánchez-Madrid, F.; Yáñez-Mo, M. CD81 regulates cell migration through its association with Rac GTPase. Mol. Biol. Cell. 2013, 24, 261–273.
- Bailly, C.; Beignet, J.; Loirand, G.; Sauzeau, V. Rac1 as a therapeutic anticancer target: Promises and limitations. Biochem. Pharmacol. 2022, 203, 115180.
- Antalíková, J.; Sečová, P.; Michalková, K.; Horovská, Ľ.; Páleníková, V.; Jankovičová, J. Expression of αV integrin and its potential partners in bull reproductive tissues, germ cells and spermatozoa. Int. J. Biol. Macromol. 2022, 209, 542–551.
- Kummer, D.; Steinbacher, T.; Thölmann, S.; Schwietzer, M.F.; Hartmann, C.; Horenkamp, S.; Demuth, S.; Peddibhotla, S.S.D.; Brinkmann, F.; Kemper, B.; et al. A JAM-A-tetraspanin-αvβ5 integrin complex regulates contact inhibition of locomotion. J. Cell. Biol. 2022, 221, e202105147.
- Oguri, Y.; Shinoda, K.; Kim, H.; Alba, D.L.; Bolus, W.R.; Wang, Q.; Brown, Z.; Pradhan, R.N.; Tajima, K.; Yoneshiro, T.; et al. CD81 Controls Beige Fat Progenitor Cell Growth and Energy Balance via FAK Signaling. Cell 2020, 182, 563–577.
- Yu, J.; Lee, C.Y.; Changou, C.A.; Cedano-Prieto, D.M.; Takada, Y.K.; Takada, Y. The CD9, CD81, and CD151 EC2 domains bind to the classical RGD-binding site of integrin αvβ3. Biochem. J. 2017, 474, 589–596.
- Gustafson-Wagner, E.; Stipp, C.S. The CD9/CD81 tetraspanin complex and tetraspanin CD151 regulate α3β1 integrin-dependent tumor cell behaviors by overlapping but distinct mechanisms. PLoS ONE 2013, 8, e61834.
- Torres-Gómez, Á.; Cardeñes, B.; Díez-Sainz, E.; Lafuente, E.M.; Cabañas, C. Functional Integrin Regulation Through Interactions with Tetraspanin CD9. Methods Mol. Biol. 2021, 2217, 47–56.
- Bruening, J.; Lasswitz, L.; Banse, P.; Kahl, S.; Marinach, C.; Vondran, F.W.; Kaderali, L.; Silvie, O.; Pietschmann, T.; Meissner, F.; et al. Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB. PLoS Pathog. 2018, 14, e1007111.
This entry is offline, you can click here to edit this entry!
