Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Cells need an antioxidant defense barrier against oxidizing molecules to establish cell hemostasis. Antioxidants are nucleophiles that have a high affinity to react with electrophilic reactive species and neutralize them. Glutathione (GSH), as the most abundant endogenous antioxidant molecule, displays a specific role in the oxidative response. It can effectively eliminate ROS and decreases the generation of the oxidative signal. In this process, GSH donates an electron to form two oxidized GSH (GSSG).
- oxidative stress
- Keap1/Nrf2 pathway
- Glutathione
- reactive oxygen species
- FOXO pathway
1. FOXO Pathways Activation and Its Consequences
FOXO transcription factors are parts of the Forkhead box family of transcription factors, including FOXO1, FOXO3a, FOXO4, and FOXO6 [1]. They are involved in various cellular functions, including cell proliferation, apoptosis, differentiation, modulation of oxidative stress, and DNA damage [2][3].
Figure 1 shows a common five-domain structure/organization of FOXO proteins, and their arrangement in the sequence has been elucidated: N-terminal region (CR1), winged-helix DNA-binding domain (DBD) known as Forkhead domain ((PDB ID 6QVW), nuclear localization sequence (NLS), nuclear export signal (NES) and C-terminal conserved regions (CR3), also known as transactivation domain (TAD) [4]. DBD consists of about 110 amino acid residues, which are folded at the secondary structure level into three α-helices (H1, H2, and H3), a three-stranded antiparallel β-sheet comprising three twisted strands (S1-S2 and S3), and two wing-like loops (L1 and L2) which are arranged H1-S1-H2-H3-S2-L1-S3-L2 sequence [3]. Among them, the helix H3, in collaboration with loops, is responsible for recognizing and specifically binding to “forkhead-responsive DNA elements” (FHRE) with a core consensus sequence 5′-(A/C)AA(C/T)A-3′ in the major groove of the DNA [5].
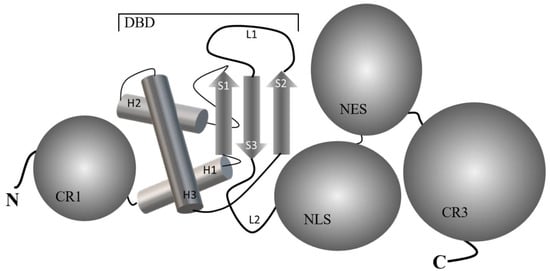
Figure 1. Common five-domain structure/organization of FOXO proteins. The graphical structure of the DNA binding domain (DBD) from FOXO1 has been presented based on the solution NMR method (PDB-ID 6QVW). H1, H2, and H3 are helices, S1, S2, and S3 are strands, and L1 and L2 are wing-like loops. CR1 and CR2 depict N-terminal and C-terminal region domains, respectively. Nuclear localization signal (NLS) and nuclear export sequence (NES) domains are also depicted.
Several pieces of evidence show that the deregulation of FOXO proteins is connected with tumorigenesis and cancer development. Based on various studies, FOXO factors are both sensors of oxidative stress signals and effectors of the consequent cellular response. FOXO proteins control the intracellular redox environment through some mechanisms. Activation of FOXO increases manganese-dependent SOD (MnSOD or SOD2) and CAT, as well as other opposing proteins against oxidative stress, such as sestrin 3 and PTEN-induced kinase 1 (PINK1) [6]. ROS activates FOXO indirectly or directly. Indirect activation operates via the phosphorylation of FOXO by activated c-Jun N-terminal kinases (JNK) and p38 MAPK cascades [7] (Figure 2). Direct FOXO modification through targeted sulfhydryl group (−SH) oxidation on FOXO to generate disulfides or persulfides [8].
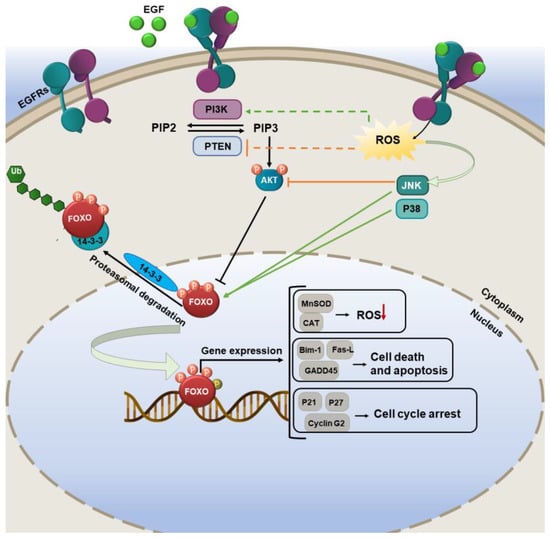
Figure 2. The FOXO pathway regulation by ROS. In oxidative conditions, phosphorylation of FOXO family members by various upstream effectors can trigger some antioxidant enzyme expression and reduce ROS levels in cells.
Thioredoxin binding protein (TXNIP) has multiple functions and plays an important role in redox homeostasis [9]. FOXO, through modulating TXNIP, can promptly activate thioredoxin toward reducing the cellular ROS levels in glucose-treated endothelial cells [10]. The interaction of FOXO1 with sirtuin 1 (SIRT1) under oxidative conditions led to the triggering of anti-stress-related genes, thus boosting the growth and survival of the cells [11]. Based on some studies, FOXO has a potent correlation with p53 in cell cycle regulation and tumor suppression [12][13]. p53 can directly target FOXO3a and increases its nuclear translocation and induction of apoptosis. FOXO can trigger the induction of autophagy in cells through the upregulation of various autophagy-related genes such as autophagy-related genes (Atg4, Atg7, and Atg14) [14][15].
2. Nrf2 Pathway Activation and Its Consequences
Nrf2, as a pivotal redox-sensitive transcription factor, controls the expression of an array of antioxidant proteins and plays a fundamental role in antioxidant response. Nrf2 contains 605 amino acids that are classified into seven efficient Nrf2-ECH homology domains (Neh1-7) [16][17] (Figure 3a). Neh1 is involved in ARE and DNA binding; moreover, Neh1 and Neh6 have the main role in regulating Nrf2 stability [18]. The Neh2 domain, at the N-terminal, comprises seven lysine residues responsible for ubiquitination, and ETGE and DLG motifs involve in Keap1 binding [19]. Interaction of Neh3, Neh4, and Neh5 with coactivators can prompt the activation of Nrf2-related genes. Finally, Neh7, through binding to retinoic X receptor α (RXRα), an Nrf2 inhibitor, can negatively regulate Nrf2-related genes [20].
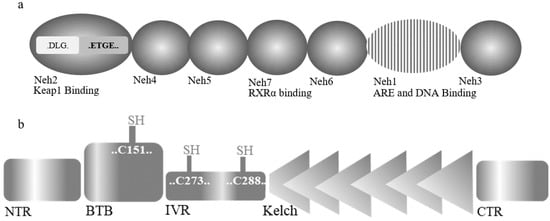
Figure 3. Domain structures of (a) Nrf2 and (b) Keap1.
Kelch-like ECH-associated protein 1 (Keap1), a zinc-metalloprotein, is the essential regulator of Nrf2 activity [21]. Keap1 protein is composed of 624 amino acids and has five structural regions: N-terminal region (NTR), common complex/tramtrack/bric-a-brac (BTB) domain, intervening region (IVR), Kelch domain, and C-terminal region (CTR) (Figure 3b). BTB domain mediates the homodimerization of Keap1 and Cullin3 (Cul3). Keap1 also has 27 cysteine residues, among them with more than 20 free sulfhydryl groups [22][23]. These highly reactive groups act as stress sensors in the oxidative condition where modification of these residues can modulate its function. Cys273 and Cys288 are the most important residues for Keap1 to regulate Nrf2 under normal and stress conditions, while Cys151 in the BTB domain is generally recognized as the most necessary sensory element under oxidative conditions [24][25][26].
In normal conditions, Keap1 binds to Nrf2 in the cytosol and, through forming a complex with cullin3 (CUL3), catalyzes the poly-ubiquitination of Nrf2 and its degradation [27]. Under oxidative stress conditions, the disassociation of Nrf2 and Keap1 and the nuclear translocation of Nrf2 leads to the upregulation of various cytoprotective genes [28]. Furthermore, modulation of Nrf2 activity can occur through the phosphorylation of several Ser/Thr residues by numerous kinases, including MAPKs, PI3K/AKT, and PKC [29]. Nrf2-related oxidative response in cells involves boosting the expression of various antioxidant enzymes such as SOD, CAT, HO-1, thioredoxin reductase (TrxR), glutathione reductase (GR), and NAD[P]H quinone dehydrogenase-1 (NQO-1) [16].
The SODs are a family of enzymes that efficiently catalyze the dismutation of the superoxide radical anion and produce hydrogen peroxide (H2O2) and H2O. Three isoforms of this enzyme comprise the cytosolic-CuZnSOD (Sod1), mitochondrial superoxide dismutase MnSOD (Sod2), and the extracellular secreted enzyme, which is attached to proteoglycans and the cell surface [30]. Exposure to H2O2 is a generally used process to trigger oxidative damage in cells. The Fenton reaction between H2O2 and Fe2+ ions increases hydroxyl radical levels and is a central mechanism for the exacerbation of oxidative damage. The peroxide formed is modified by the enzymes of the glutathione redox cycle and catalase [31].
Several studies have depicted that the activation of the thioredoxin (Trx) system comprising Trx, NADPH, and thioredoxin reductase (TrxR) in particularly TrxR1, is important for counteracting Nrf2 activation [32]. These observations are strongly suggestive of direct functional links between TrxR1 and Nrf2 [33]. In mammalian cells, there are two main thiol-dependent antioxidant systems, a 12 kDa oxidoreductase protein, thioredoxin (Trx), and the most abundant non-protein thiol-harboring molecule, glutathione (GSH). The Trx active site contains two key cysteine residues in a CXXC motif able to reversibly oxidize into a dithiol and reduce other substrate proteins as an essential component of the redox activity of the Trx system. This system, through providing electrons for various enzymes, exerts a fundamental function in the protection of DNA against oxidative stress [34].
According to previous studies, hyperactivation of Nrf2 can initiate multiple signaling pathways in the cells and is involved in proliferation, survival, metabolic reprogramming, angiogenesis, drug resistance, and metastasis [35]. Therefore, a detailed examination of the antioxidant elements in the Nrf2 pathway is very important to understand their function in cells under acute or chronic stress conditions.
HO-1 has been identified as a functional effector of Nrf2-related cell responses [36][37]. Some evidence identified that this stress protein plays a different role in normal and cancer cells. This means that some of the oncogenic activities of Nrf2 in cancer cells may be the results of the activity of this enzyme [38].
This entry is adapted from the peer-reviewed paper 10.3390/antiox12030735
References
- Tia, N.; Singh, A.K.; Pandey, P.; Azad, C.S.; Chaudhary, P.; Gambhir, I.S. Role of Forkhead Box O (FOXO) transcription factor in aging and diseases. Gene 2018, 648, 97–105.
- Wang, Y.; Zhou, Y.; Graves, D.T. FOXO transcription factors: Their clinical significance and regulation. BioMed Res. Int. 2014, 2014, 925350.
- Huang, H.; Tindall, D.J. Dynamic FoxO transcription factors. J. Cell Sci. 2007, 120, 2479–2487.
- Psenakova, K.; Kohoutova, K.; Obsilova, V.; Ausserlechner, M.J.; Veverka, V.; Obsil, T. Forkhead domains of FOXO transcription factors differ in both overall conformation and dynamics. Cells 2019, 8, 966.
- Brent, M.M.; Anand, R.; Marmorstein, R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 2008, 16, 1407–1416.
- Charitou, P.; Burgering, B.M. Forkhead box (O) in control of reactive oxygen species and genomic stability to ensure healthy lifespan. Antioxid. Redox Signal. 2013, 19, 1400–1419.
- Zhou, Y.-Y.; Li, Y.; Jiang, W.-Q.; Zhou, L.-F. MAPK/JNK signalling: A potential autophagy regulation pathway. Biosci. Rep. 2015, 35, e00199.
- Myatt, S.S.; Brosens, J.J.; Lam, E.W.-F. Sense and sensitivity: FOXO and ROS in cancer development and treatment. Antioxid. Redox Signal. 2011, 14, 675–687.
- Zhou, J.; Chng, W.-J. Roles of thioredoxin binding protein (TXNIP) in oxidative stress, apoptosis and cancer. Mitochondrion 2013, 13, 163–169.
- De Candia, P.; Blekhman, R.; Chabot, A.E.; Oshlack, A.; Gilad, Y. A combination of genomic approaches reveals the role of FOXO1a in regulating an oxidative stress response pathway. PLoS ONE 2008, 3, e1670.
- Li, D.; Ding, Z.; Du, K.; Ye, X.; Cheng, S. Reactive oxygen species as a link between antioxidant pathways and autophagy. Oxidative Med. Cell. Longev. 2021, 2021, 5583215.
- Donehower, L.A.; Harvey, M.; Slagle, B.L.; McArthur, M.J.; Montgomery, C.A.; Butel, J.S.; Bradley, A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992, 356, 215–221.
- Jacks, T.; Remington, L.; Williams, B.O.; Schmitt, E.M.; Halachmi, S.; Bronson, R.T.; Weinberg, R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994, 4, 1–7.
- Cheng, Z. The FoxO–autophagy axis in health and disease. Trends Endocrinol. Metab. 2019, 30, 658–671.
- Brown, A.K.; Webb, A.E. Regulation of FOXO factors in mammalian cells. Curr. Top. Dev. Biol. 2018, 127, 165–192.
- Ghareghomi, S.; Rahban, M.; Moosavi-Movahedi, Z.; Habibi-Rezaei, M.; Saso, L.; Moosavi-Movahedi, A.A. The potential role of curcumin in modulating the master antioxidant pathway in diabetic hypoxia-induced complications. Molecules 2021, 26, 7658.
- Keum, Y.-S.; Choi, B.Y. Molecular and chemical regulation of the Keap1-Nrf2 signaling pathway. Molecules 2014, 19, 10074–10089.
- Zhang, Y.; Gordon, G.B. A strategy for cancer prevention: Stimulation of the Nrf2-ARE signaling pathway. Mol. Cancer Ther. 2004, 3, 885–893.
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006, 281, 24756–24768.
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.D. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013, 73, 3097–3108.
- Pu, D.; Zhao, Y.; Chen, J.; Lv, A.; Zhu, S.; Luo, C.; Zhao, K.; Xiao, Q. Protective effects of sulforaphane on cognitive impairments and AD-like lesions in diabetic mice are associated with the upregulation of Nrf2 transcription activity. Neuroscience 2018, 381, 35–45.
- Kobayashi, M.; Li, L.; Iwamoto, N.; Nakajima-Takagi, Y.; Kaneko, H.; Nakayama, Y.; Eguchi, M.; Wada, Y.; Kumagai, Y.; Yamamoto, M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 2009, 29, 493–502.
- Yamamoto, T.; Suzuki, T.; Kobayashi, A.; Wakabayashi, J.; Maher, J.; Motohashi, H.; Yamamoto, M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008, 28, 2758–2770.
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003, 23, 8137–8151.
- Levonen, A.-L.; Landar, A.; Ramachandran, A.; Ceaser, E.K.; Dickinson, D.A.; Zanoni, G.; Morrow, J.D.; Darley-Usmar, V.M. Cellular mechanisms of redox cell signalling: Role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem. J. 2004, 378, 373–382.
- Wakabayashi, N.; Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Kang, M.-I.; Kobayashi, A.; Yamamoto, M.; Kensler, T.W.; Talalay, P. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 2004, 101, 2040–2045.
- Suzuki, T.; Yamamoto, M. Stress-sensing mechanisms and the physiological roles of the Keap1–Nrf2 system during cellular stress. J. Biol. Chem. 2017, 292, 16817–16824.
- Zhang, J.; Wang, X.; Vikash, V.; Ye, Q.; Wu, D.; Liu, Y.; Dong, W. ROS and ROS-mediated cellular signaling. Oxidative Med. Cell. Longev. 2016, 2016, 4350965.
- Liu, T.; Lv, Y.-F.; Zhao, J.-L.; You, Q.-D.; Jiang, Z.-Y. Regulation of Nrf2 by phosphorylation: Consequences for biological function and therapeutic implications. Free. Radic. Biol. Med. 2021, 168, 129–141.
- Miller, T.W.; Isenberg, J.S.; Roberts, D.D. Molecular regulation of tumor angiogenesis and perfusion via redox signaling. Chem. Rev. 2009, 109, 3099–3124.
- Reddy, V.; Kasahara, E.; Hiraoka, M.; Lin, L.-R.; Ho, Y.-S. Effects of variation in superoxide dismutases (SOD) on oxidative stress and apoptosis in lens epithelium. Exp. Eye Res. 2004, 79, 859–868.
- Nagarajan, N.; Oka, S.; Sadoshima, J. Modulation of signaling mechanisms in the heart by thioredoxin 1. Free. Radic. Biol. Med. 2017, 109, 125–131.
- Cebula, M.; Schmidt, E.E.; Arnér, E.S. TrxR1 as a potent regulator of the Nrf2-Keap1 response system. Antioxid. Redox Signal. 2015, 23, 823–853.
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free. Radic. Biol. Med. 2014, 66, 75–87.
- Ghareghomi, S.; Habibi-Rezaei, M.; Arese, M.; Saso, L.; Moosavi-Movahedi, A.A. Nrf2 Modulation in Breast Cancer. Biomedicines 2022, 10, 2668.
- Furfaro, A.; Traverso, N.; Domenicotti, C.; Piras, S.; Moretta, L.; Marinari, U.; Pronzato, M.; Nitti, M. The Nrf2/HO-1 axis in cancer cell growth and chemoresistance. Oxidative Med. Cell. Longev. 2016, 2016, 1958174.
- Mangano, K.; Cavalli, E.; Mammana, S.; Basile, M.S.; Caltabiano, R.; Pesce, A.; Puleo, S.; Atanasov, A.G.; Magro, G.; Nicoletti, F. Involvement of the Nrf2/HO-1/CO axis and therapeutic intervention with the CO-releasing molecule CORM-A1, in a murine model of autoimmune hepatitis. J. Cell. Physiol. 2018, 233, 4156–4165.
- Na, H.-K.; Surh, Y.-J. Oncogenic potential of Nrf2 and its principal target protein heme oxygenase-1. Free. Radic. Biol. Med. 2014, 67, 353–365.
This entry is offline, you can click here to edit this entry!
