After antibiotics are used in livestock and poultry, on the one hand, selection pressure will be formed to make the intestinal microorganisms of livestock and poultry develop resistance, thus making livestock and poultry manure carry a large amount of antibiotic resistance gene (ARG); on the other hand, about 30-90% of antibiotics will be discharged into the environment with livestock and poultry manure, and the antibiotics entering the environment will not only cause chemical pollution, but most importantly, may induce antibiotic-resistant bacteria (ARB) and ARG in the environment generation in the environment. The sources of ARGs in livestock wastewater may be threefold: (1) livestock wastewater receives ARGs already present in livestock manure; (2) pollutants such as antibiotics and heavy metals in wastewater induce microorganisms to produce ARGs; and (3) proliferation of microbial host bacteria leads to proliferation of ARGs. Unlike traditional chemical pollutants, which exhibit unique environmental behaviors such as replicability, transmissibility, and environmental persistence due to their inherent biological properties, ARGs are promoted by mobile genetic elements such as plasmids, transposons, integrons, insertion sequence common regions, and complex integrons. These ARGs are transmitted between different microorganisms in environmental media through horizontal gene transfer (HGT) mechanisms and may enter the food chain and humans through direct or indirect routes, increasing human drug resistance and endangering human public health.
- plant
- antibiotic
- root
- ARGs
1. Effectiveness of Plant Ecological Treatment Technology on the Removal of Antibiotic Resistance Genes
| Wastewater Types | Botany Types | Variable Factors |
Target ARGs | Removal Effects | References |
|---|---|---|---|---|---|
| Domestic wastewater | Cyperus alternifolius L. | Artificial aeration and mixing design | sul1, sul2, tetG, tetO, ermB, qnrS, qnrD, cmlA and floR | 87.8~99.1% | [9] |
| Domestic wastewater | Thalia dealbata Fraser. and Iris tectorum Maxim. | Flow patterns and plant types | sul1, sul2, sul3, tetG, tetM, tetO, tetX, ermB, ermC, cmlA and floR | 63.9~84.0% | [3] |
| Domestic wastewater | Cyperus alternifolius L. | Substrate and hydraulic load | sul1, sul2, sul3, tetG, tetM, tetO, tetX, ermB, ermC, qnrB, qnrD, qnrS, cmlA, fexA, fexB, floR, intl1 and intl2 | 50.0~85.8% | [5] |
| Pig farm wastewater | P. australis | Vertical Flow Artificial Wetland | sul1, sul2 and sul3 | 89%, 88% and 84% | [10] |
| Pig farm wastewater | Hybrid pennisetum | Filler type | tetM, tetO and tetW | 50% | [11] |
| Pig farm wastewater | Arundo donax | Filler type | sulI, sulII, sulIII, tetM, tetO and tetW | 67.5%, 85.6%, 95.6%, 87.9%, 97.9% and 98.5% | [4] |
| Synthetic pig farm wastewater | P. australis | Water flow method | sulI, sulII, tetM, tetW and tetO | 99.9% (Sulfonamides); 99.9% (Tetracycline) | [8] |
| Livestock wastewater | P. australis | Exogenous antibiotics and resistant bacteria | 73 ARGs | >60% | [12] |
| Pig farm wastewater after digestion | Iris pseudacorus | With or without aeration | tetA, tetM, tetO and tetW | 87.88% | [13] |
| Urban wastewater | P. australis | Operating conditions | intI1, qnrS, sul1, sul2, blaTEM and ermB | −7.67~92.9% | [2] |
| Wetlands wastewater | P. australis | With or without aeration | sul1, sul2, tetA, tetC, ermB and intl1 | 12.3~39.2% | [14] |
| Pig farm wastewater | Pontederia cordata and M. verticillatum L. | Water flow method | sul3, intI1, sul2, sul1, tetO, ermB, intI2, tetB/P, ermC, tetM and tetX | 87~99% | [15] |
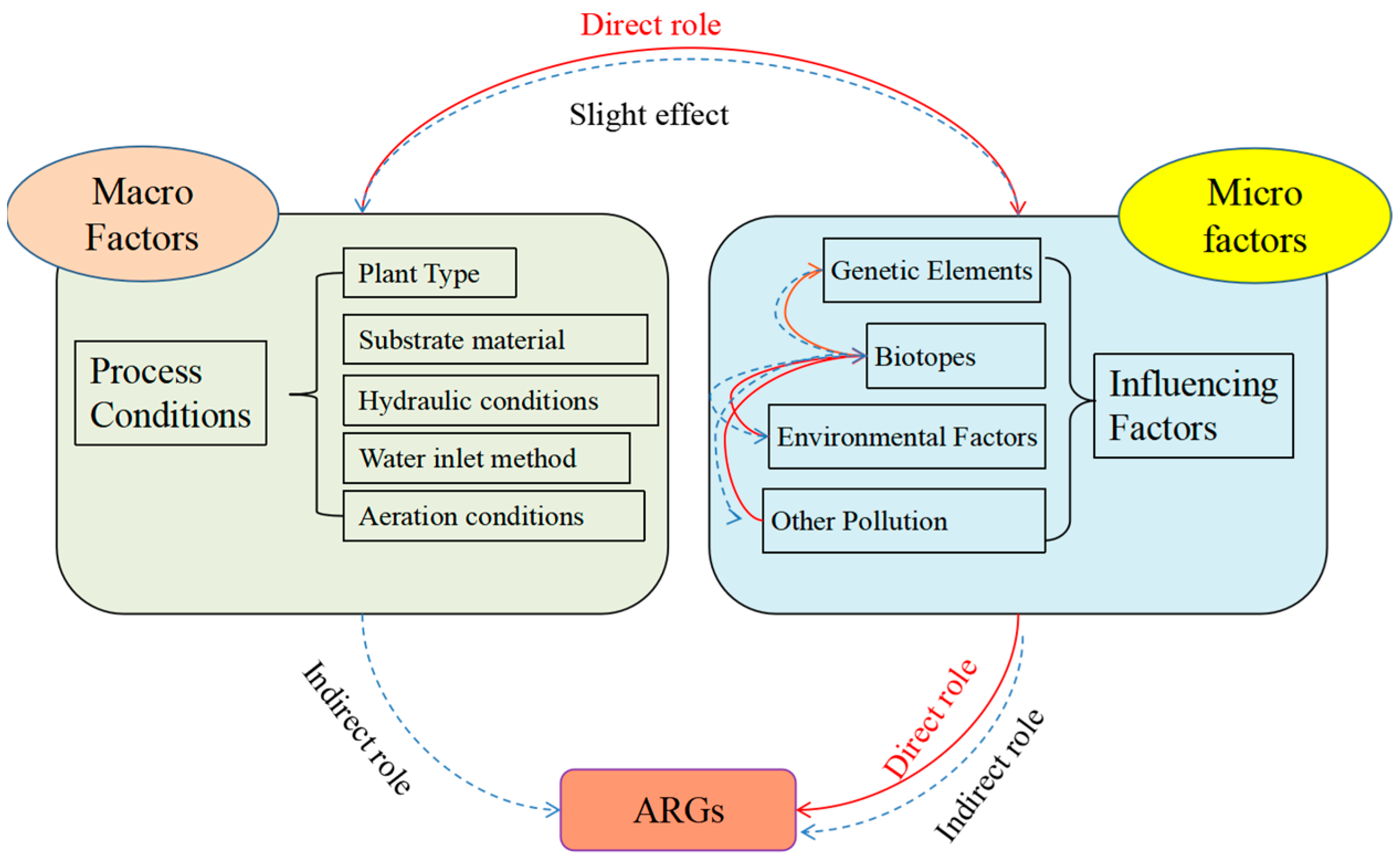
2. Drivers of Resistance Gene Elongation in Plant Ecological Treatment Systems
3. Transmission Pathways and Distribution Characteristics of Antibiotic Resistance Genes in Plant Tissues
4. Mechanism of Removal of Antibiotic Resistance Genes

This entry is adapted from the peer-reviewed paper 10.3390/ijerph20054357
References
- García, J.; García-Galán, M.J.; Day, J.W.; Boopathy, R.; White, J.R.; Wallace, S.; Hunter, R.G. A review of emerging organic contaminants (EOCs), antibiotic resistant bacteria (ARB), and antibiotic resistance genes (ARGs) in the environment: Increasing removal with wetlands and reducing environmental impacts. Bioresour. Technol. 2020, 307, 123228.
- Ávila, C.; García-Galán, M.J.; Borrego, C.M.; Rodríguez-Mozaz, S.; García, J.; Barceló, D. New insights on the combined removal of antibiotics and ARGs in urban wastewater through the use of two configurations of vertical subsurface flow constructed wetlands. Sci. Total Environ. 2021, 755, 142554.
- Chen, J.; Ying, G.G.; Wei, X.D.; Liu, Y.S.; Liu, S.S.; Hu, L.X.; He, L.Y.; Chen, Z.F.; Chen, F.R.; Yang, Y.Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Effect of flow configuration and plant species. Sci. Total Environ. 2016, 571, 974–982.
- Du, L.; Zhao, Y.; Wang, C.; Zhang, H.; Chen, Q.; Zhang, X.; Zhang, L.; Wu, J.; Wu, Z.; Zhou, Q. Removal performance of antibiotics and antibiotic resistance genes in swine wastewater by in-tegrated vertical-flow constructed wetlands with zeolite substrate. Sci. Total Environ. 2020, 721, 137765.
- Chen, J.; Wei, X.D.; Liu, Y.S.; Ying, G.G.; Liu, S.S.; He, L.Y.; Su, H.C.; Hu, L.X.; Chen, F.R.; Yang, Y.Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Optimization of wetland substrates and hydraulic loading. Sci. Total Environ. 2016, 565, 240–248.
- Huang, X.; Liu, C.; Li, K.; Liu, F.; Liao, D.; Liu, L.; Zhu, J.; Liao, J. Occurrence and distribution of veterinary antibiotics and tetracycline resistance genes in farmland soils around swine feedlots in Fujian Province, China. Environ. Sci. Pollut. Res. 2013, 20, 9066–9074.
- Gao, P.P.; Mao, D.Q.; Luo, Y.; Wang, L.; Xu, B.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aqua-culture environment. Water Res. 2012, 46, 2355–2364.
- Liu, L.; Liu, Y.; Wang, Z.; Liu, C.X.; Huang, X.; Zhu, G.F. Behavior of tetracycline and sulfamethazine with corresponding resistance genes from swine wastewater in pilot-scale constructed wetlands. J. Hazard. Mater. 2014, 278, 304–310.
- Chen, J.; Deng, W.J.; Liu, Y.S.; Hu, L.X.; He, L.Y.; Zhao, J.L.; Wang, T.T.; Ying, G.G. Fate and removal of antibiotics and antibiotic resistance genes in hybrid constructed wetlands. Environ. Pollut. 2019, 249, 894–903.
- Ma, J.; Cui, Y.; Li, A.; Zou, X.; Ma, C.; Chen, Z. Antibiotics and antibiotic resistance genes from wastewater treated in constructed wetlands. Eco-Log. Eng. 2022, 177, 106548.
- Liu, L.; Liu, C.; Zheng, J.; Huang, X.; Wang, Z.; Liu, Y.; Zhu, G. Elimination of veterinary antibiotics and antibiotic resistance genes from swine wastewater in the vertical flow constructed wetlands. Chemosphere 2013, 91, 1088–1093.
- Liu, L.; Xin, Y.; Huang, X.; Liu, X. Response of antibiotic resistance genes in constructed wetlands during treatment of livestock wastewater with different exogenous inducers: Antibiotic and antibiotic-resistant bacteria. Bioresour. Technol. 2020, 314, 123779.
- Feng, L.; Wu, H.; Zhang, J.; Brix, H. Simultaneous elimination of antibiotics resistance genes and dissolved organic matter in treatment wetlands: Characteristics and associated relationship. Chem. Eng. J. 2021, 415, 128966.
- Ma, J.; Cui, Y.; Li, A.; Zhang, W.; Liang, J.; Wang, S.; Zhang, L. Evaluation of the fate of nutrients, antibiotics, and antibiotic resistance genes in sludge treatment wetlands. Sci. Total Environ. 2020, 712, 136370.
- Chen, J.; Liu, Y.S.; Su, H.C.; Ying, G.G.; Liu, F.; Liu, S.S.; He, L.Y.; Chen, Z.F.; Yang, Y.Q.; Chen, F.R. Removal of antibiotics and antibiotic resistance genes in rural wastewater by an integrated constructed wetland. Environ. Sci. Pollution Res. 2015, 22, 1794–1803.
- Chen, C.; Xia, K. Fate of land applied emerging organic contaminants in waste materials. Curr. Pollut. Rep. 2017, 3, 38–54.
- Duan, M.; Li, H.; Gu, J.; Tuo, X.; Sun, W.; Qian, X.; Wang, X. Effects of biochar on reducing the abundance of oxytetracycline, antibiotic resistance genes, and human pathogenic bacteria in soil and lettuce. Environ. Pollut. 2017, 224, 787–795.
- Zhu, B.; Chen, Q.; Chen, S.; Zhu, Y.G. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environ. Int. 2017, 98, 152–159.
- Zhi, S.; Ding, G.; Li, A.; Guo, H.; Shang, Z.; Ding, Y.; Zhang, K. Fate of antibiotic resistance genes during high solid anaerobic digestion with pig manure: Focused on different starting modes. Bioresour. Technol. 2021, 328, 124849.
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317.
- Wang, P.; Yuan, Q.; Zhou, W. Study on photocatalytic degradation and reaction kinetics of tetracycline antibiotics in biogas slurry. Trans. Chin. Soc. Agric. Eng. 2018, 34, 193–198.
- Berg, J.; Thorsen, M.K.; Holm, P.E.; Jensen, J.; Nybroe, O.; Brandt, K.K. Cu exposure under field conditions coselects for antibiotic resistance as determined by a novel cultivation-independent bacterial community tolerance assay. Environ. Sci. Technol. 2010, 44, 8724–8728.
- Yang, Q.; Ren, S.; Niu, T.; Guo, Y.; Qi, S.; Han, X.; Liu, D.; Pan, F. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables. Environ. Sci. Pollut. Res. 2014, 21, 1231–1241.
- Ye, M.; Sun, M.; Feng, Y.; Wan, J.; Xie, S.; Tian, D.; Zhao, Y.; Wu, J.; Hu, F.; Li, H.; et al. Effect of biochar amendment on the control of soil sulfonamides, antibiotic-resistant bacteria, and gene enrichment in lettuce tissues. J. Hazard. Mater. 2016, 309, 219–227.
- Guo, X.P.; Li, J.; Yang, F.; Yang, J.; Yin, D. Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci. Total Environ. 2014, 493, 626–631.
- Yang, Y.; Li, B.; Zou, S.H.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106.
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Philipp Rauf, P.; Huettel, B.; Reinhardt, R.; Elmon Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95.
- He, S.; Wang, Y.M.; Li, C.S.; Li, Y.; Zhou, J. The nitrogen removal performance and microbial communities in a two-stage deep sequencing constructed wetland for advanced treatment of secondary effluent. Bioresour. Technol. 2018, 248, 82–88.
- He, T.; Wei, G.; Luan, Z.Y.; Xie, S.G. Spatiotemporal variation of bacterial and archaeal communities in a pilot-scale constructed wetland for surface water treatment. Appl. Microbiol. Biotechnol. 2015, 100, 1479–1488.
- Chen, Q.L.; An, X.L.; Zhu, Y.G.; Xie, S. Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 2017, 51, 8149–8157.
- Duran, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 2018, 175, 973–983.
- Zhang, Y.J.; Hu, H.W.; Chen, Q.L.; Singh, B.K.; Yan, H.; Chen, D.; He, J.Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019, 130, 104912.
- Frank, A.; Saldierna, G.J.; Shay, J. Transmission of bacterial endophyte. Microorganisms 2017, 5, 70.
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206.
