Understanding the packaging of DNA into chromatin is essential for the study of gene expression regulatory mechanisms. Heterochromatin establishment and maintenance dynamics have emerged as key features involved in genome stability, cellular growth, and disease. The heterochromatin protein HP1a is the most extensively studied factor that has both establishment and heterochromatin maintenance activities. This protein has two primary domains, namely the chromoshadow and the chromodomain, separated by a hinge region. Several works have taken place over the years, taking the challenge of defining HP1a partners using diverse experimental approaches. We revised and assemble on explaining these interactions and the potential complexes and subcomplexes associated formed with this essential protein. Characterization of these complexes will allow us to clearly understand the consequences of HP1a interactions in heterochromatin in maintenance, heterochromatin dynamics, and the direct relationship of heterochromatin with gene regulation.
- HP1a, Heterochromatin, Drosophila
1. Introduction
Chromatin is a complex of DNA and associated proteins in which the genetic material is packed in the interior of the nucleus of eukaryotic cells [1]. To organize this highly compact structure, two categories of proteins are needed: histones [2] and accessory proteins, such as chromatin regulators and histone-modifying proteins. Both kinds of proteins participate in maintaining the structure of chromatin and regulating gene expression [3]. The primary unit of chromatin is the nucleosome [4], which is formed by an octamer of histones, with two copies of histones H2A, H2B, H3, and H4 (also called the canonical or core histones) [4]. The histone H1 has been referred to as a “linker” because a single copy is positioned on the DNA between each nucleosome [5]. Deciphering the procedures that control chromatin packaging has become a significant issue in understanding developmental programs and disease states.
There are two primary types of chromatin in the nucleus: heterochromatin and euchromatin [6]. Heterochromatin is abundant in compacted, highly condensed, silenced, and repetitious sequences found near centromeric and telomeric locations. By contrast, euchromatin includes the majority of transcriptionally active genes [7]. Through staining different types of cells, Emil Heitz conceived the term “heterochromatin” more than 90 years ago, observing retention of this more compact structure throughout the cell cycle [8]. These core heterochromatic structures have become an essential area of study because of their role in gene silencing [9].
In all eukaryotes, constitutive heterochromatin is established early in development. During the 1960s, satellite sequences were identified, sequenced, and mapped to pericentromeric and telomeric regions of metaphase chromosomes located at the nuclear periphery of interphase cells [10]. With the development of automatic sequencing over the decades that followed, studies on vertebrates have determined that the genome is rich in repetitive sequences that, for example, account for more than 50% of the human genome. There are many types of these repetitive elements: some are composed of retrotransposon sequences, others of long and short interspersed elements known as LINEs, SINEs, Alu sequences, in addition to minor and major satellite sequences. These sequences need to be silenced to avoid chromosome instability, and several mechanisms cooperate toward maintaining this silencing. These mechanisms include DNA methylation, histone post-transcriptional modifications, histone deacetylation, binding of chromatin proteins, and non-coding RNA and RNA interference pathways [11][12][13]. Embryonic stem cells have, in general, less heterochromatin than differentiated cells. This characteristic confers plasticity. As differentiation advances, cells gain heterochromatin. Disruption of any of these heterochromatin maintenance mechanisms leads to chromosome instability and can sometimes lead to diseases such as cancer.
The mechanisms of heterochromatin formation and maintenance have been highly conserved throughout the evolution of eukaryotic cells, and understanding these mechanisms using less complex animal models has helped us to advance understanding in this important field.
Based on cytological criteria, one-third of the Drosophila melanogaster genome, including the telomeres, pericentric regions, and chromosome 4, is considered as the heterochromatin [14]. As development and differentiation progress, regions regarded as heterochromatin become more abundant as differentiated cells undergo heterochromatinization to promote gene repression and prevent inappropriate gene expression. One mechanism for achieving this is for cells to anchor chromatin to the nuclear lamina resulting in gene inactivation [15]; alternatively, the heterochromatin/euchromatin borders may be defined [16], for example, by changing the profile of chromatin as differentiation progresses, i.e., as stem cells differentiate into the mature cell type [17].
The primary mechanism used to maintain differential expression patterns is the silencing of genes, which involves packaging them in structures inaccessible to DNA-binding proteins [18]. The silencing of a specific gene or chromosomal region requires covalent modification by enzymes or complexes harboring subunits that recognize these modifications and facilitate their physical association with histones [19] and their extension throughout the chromatin fiber, creating a compacted structure (heterochromatin) which is generally believed to be inaccessible to transcription-promoting factors [20]. Heterochromatinization then becomes one of the primary mechanisms used to silence chromosomal regions.
In 1930, experiments using X-ray treatment of flies have shown that genes that were translocated from euchromatic regions to the vicinity of pericentric heterochromatin, acquired a motley pattern of expression [21]. This effect, which is caused by the repressive properties of heterochromatin, was called position effect variegation (PEV) and has been exploited from the 1980s onward for the systematic examination of factors that regulate heterochromatin formation. One of the proteins identified through this screening is heterochromatin protein 1 (HP1). It is a highly conserved protein [22] that was initially discovered in Drosophila by the group of Grigliatti in a study in which the authors found more than 50 loci that acted as suppressors of PEV. The authors identified that the protein encoded by the Su(var)2-5 locus works as a dosage-dependent modifier of PEV [23]. Since then, various studies have shown that this protein is essential for the establishment and maintenance of heterochromatin.
HP1 proteins are conserved in a variety of organisms, including fission yeast (as Swi6 and Chp2) [24][25] and also vertebrates such as amphibians (e.g., frog (xHP1α and xHP1γ)) [26], birds (e.g., chicken (HP1α, HP1β, and HP1γ)) [27], and mammals (such as mice (HP1α, HP1β, and HP1γ)) [28]. Various functions have been described for each member of the family throughout the life cycle of a cell: heterochromatin formation and maintenance, gene silencing, telomere capping, DNA repair, and control of gene expression [14]. Mutations that affect HP1 protein activities have a significant impact on organism development. For example, in Drosophila, null mutants for HP1a are lethal at the embryonic stage [29]. Although the HP1 isoforms are very similar structurally, they have different functions, and null mutants for HP1a cannot be rescued by HP1b or HP1c. Thus, HP1 proteins have been revealed to interact with a wide variety of proteins, forming different complexes [30][31][32].
2. HP1a Interaction with Insulator and Architectural Proteins
Insulators were first defined as regulatory elements that maintain the correct separation of different gene domains, thereby preventing enhancer–promoter communication and/or blocking the expansion of heterochromatin silencing. They also mediate intra- and interchromosomal interactions, which are involved in the large-scale organization of the genome [33][34].
Since the 1980s, it has been known that there are DNA sequences that delimit and isolate a region of chromatin in the Drosophila heat shock locus [35][36]. Since then, many of these sequences and the factors that bind to them have been characterized [37][38]. CTCF (CCCTC binding factor) was initially identified as a repressor capable of binding to promoters in chicken and mammalian MYC genes [39]. CTCF was later shown to have an insulator function because it indirectly regulates gene expression by preventing binding between promoters and enhancers or nearby silencers, thus avoiding the inappropriate activation or silencing of certain genes [40]. Recent advances in Hi-C technique have shown that CTCF can mediate the interactions between the boundaries of topologically associating domains (TADs) resulting in the formation of chromatin loops [41]. It should be noted that not all TADs are flanked by CTCF [42].
Although it is not known exactly how CTCF assists in loop formation, a “loop extrusion” model has been proposed. This model suggests that cohesin, which is composed of SMC proteins (structural maintenance of chromosomes) and Rad21 (which is an ortholog of Drosophila verthandi (vtd)) are directed to the chromatin with the help of the NIPBL protein. Together they “pull” the DNA strand until the cohesin ring is blocked with CTCF [43]. It is currently unclear whether the same mechanism operates in Drosophila. However, ChIP-seq experiments have identified several architectural proteins (APs) which are co-localized with CTCF at several sites in the genome.
APs are characterized by promoting contacts between regulatory elements through the formation of loops; thus, they have a role in determining the organization and architecture of chromatin [44][45]. Furthermore, it has been shown that APs can contribute to the establishment of TADs [46]. Some of them include suppressor of hairy-wing (Su(Hw)) [47], dCTCF, the D. melanogaster ortholog of mammalian CCCTC-binding factor [48], Boundary Element Associated Factors (BEAF-32A and B), GAGA Associated Factor (GAF) [49], and Zeste-white 5 (Zw5) [50]. Others that have recently been identified include Elba (made up of 3 proteins), Elba1, Elba2, and Elba3 [51], Pita, the zinc-finger protein interacting with CP190 (ZIPIC), Clamp [52], and Ibf-1 and 2 [53].
Typically, the proteins bound to the insulator sequences are necessary but not sufficient for the activity of insulators. Several cofactors are also required to establish physical contacts and anchor them to nuclear structures. In Drosophila, proteins such as the modifier of mdg4 (Mod (mdg4)) [54][55] and centrosomal protein of 190kDa (CP190) [56]; cohesins (like Rad21/Vtd); and condensins (like Cap-H2) are present in different combinations in all types of insulators and fulfill these functions [57].
The degenerate motif begins at amino acid 752, and the crotonase domain extends from amino acid 675 to 778. This suggests that HIPP1 interactions with HP1a could affect (negatively or positively) its crotonase activity to some extent. Moreover, the crotonase domain seems to have a role in the interaction of HIPP1 with Su (Hw). Presently, there are no data on whether crotonase activity could be affected by interactions with these proteins. This is an important question to address experimentally in the future.
Table 1. Architectural proteins of Drosophila and motifs for possible interaction with HP1a protein.
| Protein | PxVxL | CxVxL | LxVxL |
|---|---|---|---|
| CTCF | |||
| Su(Hw) | |||
| BEAF-32 | |||
| pita | |||
| ZIPIC | |||
| Ibf1 | |||
| Ibf2 | |||
| Mod(mdg4) | |||
| CP190 | |||
| Cap-H2 | X | ||
| Elba1 | |||
| Elba2 | |||
| Elba3 | X | ||
| Shep | |||
| Zw5 | X | ||
| Clamp | |||
| GAF | X | ||
| Nip-b | |||
| Vtd | X | ||
| SA | X | ||
| Smc1 | |||
| Smc2 | X | ||
| Smc3 | |||
| HIPP1 | X |
Since HIPP1 contains possible motifs for direct interactions with HP1a, we examined whether such motifs are also present in other APs. The results are summarized in Table 1. The cohesin complex is an important factor in maintaining the structure of chromosomes. In mammals, the cohesin complex co-localizes with CTCF throughout the genome. In many of these sites, CTCF performs its function of enhancer-blocking [58][59]. In Drosophila, there is no co-localization observed between CTCF and the cohesin complex [60], but cohesins do co-localize with other APs, such as CP190 [193]. Through a ChIP-chip analysis, it was determined that Nipped-B and cohesins are located preferably in active sites and are absent from the silenced sites [61].
The Cap-H2 protein has an LxVxL binding motif beginning at 159 aa (N-terminal); also, in Stromalin (SA), this domain is present in the middle of the protein, and the same domain was found in Smc2 in the C-terminal. In Elba3, the canonical binding site, PxVxL, is found at 156 aa and for GAF, this canonical motif is present in the N-terminal. Finally, Zw5 has a putative binding domain, CxVxL, at 449 aa, the most C-terminal part of the protein (Table 1). This opens the door to possible interactions of HP1a with architectural complexes which, in the future, would be interesting to address experimentally.
To better understand whether HP1a is co-localized with APs, we analyzed previously published modENCODE chromatin immunoprecipitation (ChIP-seq) data in Drosophila for the region spanning Abd-A and Abd-B loci (Figure 1). This region has very complicated regulation, and the insulator function is essential for the correct expression of genes within this region [62][63][64] such that the active or silenced state of one domain does not extend to an adjacent one. ChIP assays detected the presence of APs such as CP190 or CTCF, not only at the border elements but also at the promoters of genes such as Abd-b [65][66]. The interactions between protein insulators, on the one hand, manage to form loops that leave out entire domains. On the other hand, elements in the domain are brought in close contact with the gene promoter, thereby mediating correct gene expression [67][68][69].
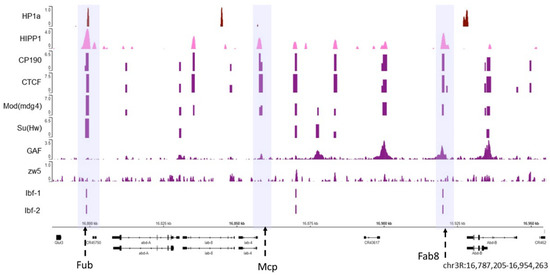
Figure 1. The HP1a and HIPP1 proteins co-localize at homeotic genes along with the AP. The HP1a and HIPP1 proteins co-localize at the Fub insulator (the first violet box) in the Abd-a gene along with CP190, Su (Hw), CTCF, and Mod (mdg4), and Ibf 1 and 2. From the previously published ChIP data on S2 cells, we find HP1a (dark red), HIPP1 (pink), and some architecture proteins (purple). The adjacent insulators Mcp and Fab8 do not co-localize with HP1a (center and right violet boxes). The regions with insulators are marked with dotted arrows inside a violet shadow. At the bottom are the Abd-a and Abd-b genes and their locations; the reference in kilobases.
As shown in Figure 1, both HP1a (dark red) and HIPP1 (pink) are present at the Fub insulator where APs, such as CP190, Su (Hw), CTCF, and Mod (mdg4) are also observed (the violet box where the Fub insulator region is present). Other known insulators are Mcp and Fab8 (highlighted in the middle and right violet boxes) [70], where APs can be observed but do not co-localize with HP1a. Moreover, not all architectural proteins are present in all the insulator loci at the same time. The Abd-b gene is silenced in S2 cells, and HP1a appears enriched at the Abd-b promoter and is flanked by APs.
The HP1a CSD domain potentially mediates all the interactions with proteins that were evaluated. This indicates that binding possibly occurs with homodimers or heterodimers of HP1a at specific regions of chromatin. The regions where we assessed the presence of architectural proteins were not constitutive heterochromatin; rather, they represent islands of facultative heterochromatin in the euchromatin. Thus, a disruption of heterochromatin may take place, where HP1a dimers cannot be formed. Subsequently, the binding of HP1a with HMTases could be impaired, which would prevent the spreading of heterochromatinization. Chromatin insulators are essential components of genome architecture across eukaryotes [71][72]. It seems plausible that HIPP1, Vtd, Nip-b, and HP1a cooperate to maintain the insulating complexes and define edges of loops, thereby facilitating the correct separation of heterochromatin and euchromatin.
3. HP1a Interaction Partners in Silenced Chromatin
Constitutive heterochromatin represents a substantial fraction of eukaryotic genomes; it plays an important role in the maintenance of genome stability and silencing of repetitive elements. Nonetheless, further studies are needed to fully understand its formation and maintenance throughout development and cell differentiation. Thorough localization studies of HP1a in Drosophila and mammals have shown that HP1a proteins associate with regions of constitutive heterochromatin around the centromeres and at the telomeres which are rich in repetitive DNA sequences. For example, in polytene chromosomes of Drosophila, mainly the chromocenter (i.e., regions of pericentric chromatin) and the telomeres are stained with HP1 antibodies [22].
Constitutive heterochromatin is established early in development. In Drosophila, it starts during MBT (in cycle 13) [73][74]. The proposed model involves a complex that contains a methyltransferase (Eggless/SetDB1) of histone H3 lysine 9 and HP1a. The histone mark H3K9me (di or tri methylated) acts as a binding site for HP1a, which binds through its CHD to these chromatin marks, possibly with the involvement of other stabilizing interactions. It is known that HP1a crosslinks nucleosomes which form condensed heterochromatic structures. For example, in yeast, HP1a also strengthens the association of the HMTase SUV39H1 to chromatin [49]. SUV39H1 methylates nearby unmethylated H3 tails at lysine 9 via its SET domain, creating new H3K9me-binding sites for HP1a. Thus, this three-component system could explain the spreading and maintenance of heterochromatic gene silencing [75][76].
Several groups have carried out chromosomal rearrangement experiments where a euchromatic gene was translocated to a heterochromatic environment and, as a result of being present in this environment, became silenced with the help of several factors, mainly HP1a [77][78]. Subsequently, experiments were carried out to direct HP1a to euchromatin regions, such as region 31 of the Drosophila 2L arm. Three of the four studied genes within this region were silenced by HP1a and the methyltransferase Su(var)3-9 [79]. These studies demonstrated HP1a to be an essential protein that promotes heterochromatin formation and gene silencing.
Different methyltransferases can work in conjunction with HP1a. For example, in null HP1 mutants, localization of Su(var)3-9 is no longer limited to the chromocenter but spreads across the chromosomes. Studies using mutants suggest that there is a sequential order in which interactions are established [80]. Another member of this complex is the zinc-finger protein Su (var) 3-7, which appears to function as an effector downstream of Su (var) 3-9 and HP1a. This protein has heterochromatic localization, very similar to that of HP1a on polytene chromosomes in pericentric regions and on chromosome 4. In addition to decreasing the dose of this protein, it reduces PEV [81]. Increasing the quantity of the product of Su (var) 3-7 prompts heterochromatin extension and epigenetic gene silencing [82]. The formation of heterochromatin is a critical developmental process. Su (var) 3-3, whose homolog in mammals is LSD1, removes H3K4me1/2 marks in early embryonic development. This led to the establishment of a balance between demethylase and methyltransferase Su (var) 3-9, contributing to the maintenance of heterochromatic domains.
The heterochromatin–euchromatin borders have previously been described cytologically [83][84] and, later, with ChIP-array analysis of genome distribution of H3K9me2 mark, and were named the “epigenomic borders” [16]. Interestingly, the epigenomic borders varied in different cell lines or tissues studied which lead authors to propose that additional mechanisms besides sequence-specific binding can establish these borders [16]. To identify the borders of pericentric heterochromatin domains more precisely, we analyzed publicly available ChIP-seq profiles in S2 cells for HP1a, along with Su (var) 3-9, Su (var) 3-7, and H3K9me3 (Figure 2, see also Material and Methods). We examined a section near the chromocenter (the black circle at the top of thde schematic representation). A clear enrichment of HP1a along with the other examined proteins and histone marks is seen in the pericentromeric region highlighted with a red rectangle. Further from the centromere (7.3 Mb), this enrichment sharply declines thus indicating the border between heterochromatin and euchromatin. APs such as CTCF and CP190 are clearly enriched just after the border in the euchromatin which is consistent with the function of these proteins to keep chromatin domains isolated from each other [85][86] and of CP190 to mark active promoters in Drosophila. Therefore, APs may play a role in defining this border. Thus, HP1a can cooperate with other factors at these epigenomic borders to maintain a correct chromatin structure. Interestingly, H3K9 acetylation is still present within the beginning of constitutive heterochromatin, co-localizing with CP190 and some CTCF peaks. These bivalent signatures may facilitate pericentromeric gene transcription, as was observed for some genes [87]. Throughout the chromosome, other HP1a sites co-localize with H3K9ac (blue shaded box, Figure 2). The HP1a enrichment sets the epigenomic border for 2R chromosome arm at 7.3 Mb position, while the epigenomic border described in [16] was set at 7.4 Mb (highlighted red square in Figure 2).
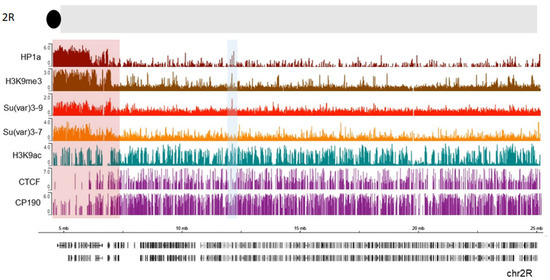
Figure 2. HP1a, together with other proteins, delimits epigenomic borders. Previously published ChIP data on S2 cells were used, where we see HP1a (dark red), Su(var) 3-9 (light red), Su(var) 3-7 (orange), H3k9me3 mark (brown), H3k9ac (green), CTCF, and CP190 (purple). The regions with a pericentric border are marked with a red rectangle according to Riddle et al. The co-localization of HP1a with H3K9ac mark is shaded in blue.
These results show that dADD1 proteins are regulators of HP1a, likely maintaining the correct local concentration of HP1a oligomers at certain regions, such as the telomeres and pericentric heterochromatin. The over- and underexpression of dADD1 can disturb the concentration of HP1a and likely affect phase transition, which could lead to chromatin instability and alterations in gene expression [88].
Moreover, it has been demonstrated that human HP1α promotes phase separation of heterochromatin from euchromatin, which is also exhibited by the Drosophila ortholog [89]. The HP1 proteins possibly involved in orchestrating these separations and play an important role in defining their possible environments and interactions. For example, HP1a could be enriched at heterochromatin regions together with a methyltransferase (as with dADD1), forming gel-type droplets with a specific environment. Interestingly, dADD1a possesses intrinsically disordered regions in the C-terminal region (650–696, 716–763, 791–1069, 1112–1132, and 1154–1199 aa) that can contribute to phase separation [90].
The contribution of HP1a to the maintenance of telomeric heterochromatin works via two main functions: as part of the CAP along with proteins such as HOAP (cav) and the repression of telomeric retrotransposons in cooperation with piRNAs [91]. Notably, HP1a localization at this heterochromatic site does not depend on its chromodomain but rather on its interaction with dADD1a in somatic cells. The interaction of HP1a and dADD1 at the telomeric region seems to be conserved because ATRX also co-localizes with HP1α at the telomeres in human cells. HP1a’s functions at telomeres seem to depend on the interactions of different proteins and even RNAs. In mammalian embryonic stem cells, ATRX (Alpha-Thalassemia with mental Retardation X-related) [92] has been shown to complex with TRIM28 and SETDB1 recruited by H3K9me3 in retrotransposon regions [93][94]. We have described the interaction of the helicase part of ATRX (XNP) together with the ADD domain (dADD1) in Drosophila in conjunction with HP1a. Furthermore, Kuroda’s laboratory was able to immunoprecipitate EGG/dSETDB1 and the Bonus protein (Trim28 homolog) together with dADD1. These proteins could form a complex and perform a similar function to their mammalian counterparts at retrotransposon regions in Drosophila.
4. HP1a Interaction Partners in Euchromatin
It is well known that HP1a does not only localize to regions of constitutive heterochromatin. A fraction of HP1a is also present within euchromatic regions of the chromosomes. For example, in polytene chromosomes of Drosophila, HP1a is found at about 200 sites within the chromosome arms. Moreover, using DamID technique numerous HP1a binding regions within euchromatic parts of Drosophila chromosome arms from various non-polytene tissues were revealed [95][96]. This points to the repressive action of HP1 within euchromatin, an interpretation supported by studies demonstrating the recruitment of HP1a by different transcriptional repressors and the reported upregulation of some HP1-bound genes in euchromatin upon mutation of HP1a in Drosophila.
Numerous experimental data demonstrate that HP1 by itself can induce heterochromatic structures and may, in fact, directly stimulate gene silencing within euchromatin. When HP1 binds to the euchromatin regions of Drosophila chromosomes through an ectopic binding domain, this process is almost always sufficient to enable the establishment of heterochromatin and silence neighboring reporter genes [97]. Furthermore, redirecting HP1α or HP1a through a GAL/lacR system to euchromatic regions in mammalian or Drosophila cells causes local condensation of higher-order chromatin structure and gene repression [3]. These experiments suggest that HP1 could indeed play a role in gene repression within euchromatic regions of chromosomes. The participation of HP1 in the regulation of euchromatic regions is even more complex and goes beyond its well-established role in gene silencing.
Unexpectedly, at some euchromatic loci, HP1 association clearly corresponds to the elevated gene expression. For example, HP1a is recruited to some of the ecdysone-induced or heat shock-induced puffs of Drosophila polytene chromosomes, generated due to strong decondensation of chromatin linked to gene activation [98], implying the modulating role of HP1a in their expression. The association of HP1 with such sites seems to be RNA-dependent since HP1 association with Hsp70 heat shock locus is only observed in the presence of RNA [98]. We also observed dADD1 enrichment at the Hsp70 locus under native conditions [98] (Figure 3). When heat shock is induced, this area becomes free of nucleosomes, the poised RNA pol II begins to elongate which results in rapid activation of transcription [99]. Another group also analyzed the presence of Xnp (the helicase part that completes the mammalian ortholog ATRX) at this locus along with Hira and the H3.3 histone variant. The authors proposed that Xnp could recognize nucleosome-free sites and work to avoid transcriptional problems or defects in subsequent DNA repair [100]. Many questions remain to be answered regarding the role of these proteins at this locus. For example, whether they strengthen the ability of HP1a to maintain a silenced state or if their presence is necessary to promote rapid transcription upon cell insult.
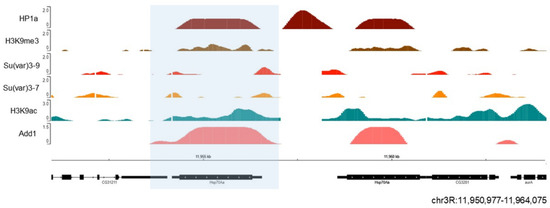
Figure 3. HP1a and dADD1 proteins co-localize at the Hsp70 locus. Previously published ChIP-seq data on S2 cells were used, where we see HP1a (dark red), dADD1 (salmon) HP1a, Su (var) 3-9 (light red), Su (var) 3-7 (orange), H3k9me3 mark (brown), and H3k9ac (green). At the bottom are the gene Hsp70 Aa (blue box) and its location, with the reference in kilobases.
Furthermore, some genes located within pericentric heterochromatin require a heterochromatic environment for their normal expression. The mutations of HP1a reduce the expression of light and rolled genes, the first to be described [101]. These genes are essential for the organism and reside naturally in heterochromatin blocks on chromosome 2. Later studies observed that a complex of HP1a and Su(var)3-9 generates very compact chromatin in these blocks [102].
High-resolution mapping of the HP1a-binding sites in Drosophila shows that HP1 is excluded from the promoter and is restricted to the transcribed regions of actively expressed genes [95]. Furthermore, HP1a depletion causes downregulation of a subset of active genes [103]. The interaction of HP1 with RNA may mediate the association of HP1 at euchromatic regions within the genome. HP1a interactions with RNA, most likely in collaboration with other interactions, recruit HP1a to these sites, where it plays a role in the promotion of gene expression [7]. This is supported by studies indicating that the section of HP1a responsible for RNA binding is the hinge [26].
It has been observed that HP1b and HP1c isoforms are more localized in the euchromatic chromosome arms than HP1a [104][105]. Lee et al. have shown that all HP1 isoforms interact with the phosphorylated at serine 2 or 5 RNA pol II, but do so with different affinities. Various localization patterns of HP1 isoforms on chromosomes may be mediated by different complexes in which these isoforms are involved (Figure 4).
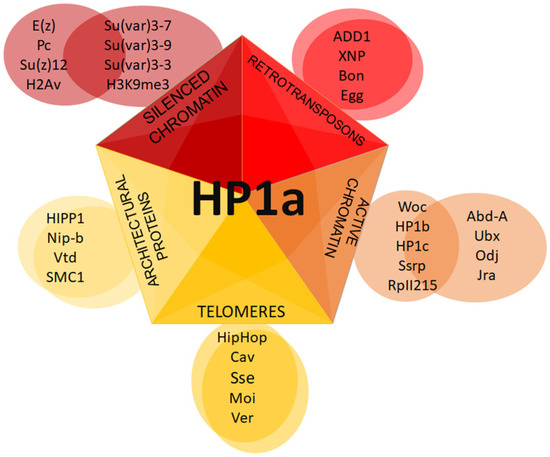
Figure 4. Putative HP1a complexes at different chromatin regions. Schematic representation of distinct complexes formed with HP1a.
5. Future Directions
Although there have been detailed genetic and biochemical studies of HP1′s roles in heterochromatin establishment and maintenance, its position at euchromatic regions and in association with RNA has not been thoroughly characterized. Valuable studies have shed light on the multiple proteins that interact with HP1a. In this extended review, we addressed whether the proteins found in association with HP1a could bear putative motifs that allow direct interaction with HP1 proteins. The identification of HP1a interactors at different chromatin regions is essential to understand the different roles of these protein complexes. We found that among the reported interactors, only a handful conserve motifs for CSD domain interactions. Experimental studies to test if these motifs function in vivo in binding to CSD would be of great importance and could extend our understanding of the biological significance of the interactions.
We also raise the possibility of HP1 interactions with architectural proteins. Indeed, we found that several architectural proteins harbor conserved putative HP1-interacting motifs (Table 1). Further experimentation will be required to understand the role of HP1a in conjunction with architectural proteins and their possible cooperation in the organization of chromatin structure [106].
Very recently, it has been shown that pericentric heterochromatin also establishes well-defined territories through contact with different proteins via the H3K9me2 mark. Most importantly, the maintenance of these territories and the established pericentromeric contacts also influence active genome regions. The protein complexes associated with these domains could also have an essential role in the formation of higher-order chromatin structures [107].
This entry is adapted from the peer-reviewed paper 10.3390/cells9081866
References
- Felsenfeld, G.; Groudine, M. Controlling the double helix. Nature 2003, 42, 448–453.
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New insights into nucleosome and chromatin structure: An ordered state or a disordered affair?. Nat. Rev. Mol. Cell Biol. 2012, 13, 436–447.
- Brueckner, L.; van Arensbergen, J.; Akhtar, W.; Pagie, L.; van Steensel, B. High-throughput assessment of context-dependent effects of chromatin proteins. Epigenetics Chromatin. 2016, 9, 1–17.
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome resolution core particle at 2.8 A. Nature 1997, 389, 251–260.
- Bayona-Feliu, A.; Casas-Lamesa, A.; Reina, O.; Bernués, J.; Azorín, F. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat. Commun. 2017, 8, 1.
- Richards, E.J.; Elgin, S.C.R. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell 2002, 108, 489–500.
- Piacentini, L.; Fanti, L.; Negri, R.; Del Vescovo, V.; Fatica, A.; Altieri, F.; Pimpinelli, S. Heterochromatin Protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009, 5, 10.
- Passarge, E. Emil Heitz and the concept of heterochromatin: Longitudinal chromosome differentiation was recognized fifty years ago. Am. J. Hum. Genet. 1979, 31, 106–115.
- Dillon, N. Heterochromatin structure and function. Biol. Cell 2004, 96, 631–637.
- Britten, R.J.; Kohne, D.E. Repeated sequences in DNA. Science 1968, 161, 529–540.
- Fodor, B.D.; Shukeir, N.; Reuter, G.; Jenuwein, T. Mammalian Su (var) genes in chromatin control. Annu. Rev. Cell Dev. Biol. 2010, 26, 471–501.
- Guthmann, M.; Burton, A.; Torres-Padilla, M. Expression and phase separation potential of heterochromatin proteins during early mouse development. EMBO Rep. 2019, 20, 1–11.
- Nishibuchi, G.; Déjardin, J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosom. Res. 2017, 25, 77–87.
- Fanti, L.; Pimpinelli, S. HP1: A functionally multifaceted protein. Curr. Opin. Genet. Dev. 2008, 18, 169–174.
- Pindyurin, A.V.; Ilyin, A.A.; Ivankin, A.V.; Tselebrovsky, M.V.; Nenasheva, V.V.; Mikhaleva, E.A.; Pagie, L.; van Steensel, B.; Shevelyov, Y.Y. The large fraction of heterochromatin in Drosophila neurons is bound by both B-type lamin and HP1a. Epigenetics Chromatin. 2018, 11, 1–17.
- Riddle, N.C.; Minoda, A.; Kharchenko, P.V.; Alekseyenko, A.A.; Schwartz, Y.B.; Tolstorukov, M.Y.; Gorchakov, A.A.; Jaffe, J.D.; Kennedy, C.; Linder-Basso, D.; et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011, 21, 147–163.
- Marshall, O.J.; Brand, A.H. Chromatin state changes during neural development revealed by in vivo cell-type specific profiling. Nat. Commun. 2017, 8, 1–9.
- Grewal, S. Heterochromatin and epigenetic control of gene expression. Science 2003, 301, 798–802.
- Wang, X.; Moazed, D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science 2017, 91, 88–91.
- Riddle, N.C.; Jung, Y.L.; Gu, T.; Alekseyenko, A.A.; Asker, D.; Gui, H.; Kharchenko, P.V.; Minoda, A.; Plachetka, A.; Schwartz, Y.B.; et al. Enrichment of HP1a on drosophila chromosome 4 genes creates an alternate chromatin structure critical for regulation in this heterochromatic domain. PLoS Genet. 2012, 8, 9.
- Muller, H.J.; Altenburg, E. The frequency of translocations produced by X-rays in Drosophila. Genetics 1930, 15, 283–311.
- James, T.C.; Elgin, S.C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 1986, 6, 3862–3872.
- Sinclair, D.A.R.; Mottus, R.C.; Grigliatti, T.A. Genes which suppress position-effect variegation in Drosophila melanogaster are clustered. MGG Mol. Gen. Genet. 1983, 191, 326–333.
- Lorentz, A.; Ostermann, K.; Fleck, O.; Schmidt, H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast , encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene 1994, 143, 139–143.
- Thon, G.; Verhein-hansen, J. Four chromo-domain proteins of schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 2000, 155, 551–568.
- Meehan, R.R.; Kao, C.F.; Pennings, S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 2003, 22, 3164–3174.
- Istomina, N.E.; Shushanov, S.S.; Springhetti, E.M.; Karpov, V.L.; Krasheninnikov, I.A.; Stevens, K.; Zaret, K.S.; Singh, P.B.; Grigoryev, S.A. Insulation of the chicken beta-globin chromosomal domain from a chromatin-condensing protein, MENT. Mol. Cell. Biol. 2003, 23, 6455–6468.
- Wreggett, K.A.; Hill, F.; James, P.S.; Hutchings, A.; Butcher, G.W.; Singh, P.B. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell Genet. 1994, 66, 99–103.
- Cowell, I.G.; Aucott, R.; Mahadevaiah, S.K.; Burgoyne, P.S.; Huskisson, N.; Bongiorni, S.; Prantera, G.; Fanti, L.; Pimpinelli, S.; Wu, R.; et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 2002, 111, 22–36.
- Pak, D.T.; Pflumm, M.; Chesnokov, I.; Huang, D.W.; Kellum, R.; Marr, J.; Romanowski, P.; Botchan, M.R. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 1997, 91, 311–323.
- Polioudaki, H.; Kourmouli, N.; Drosou, V.; Bakou, A.; Theodoropoulos, P.A.; Singh, P.B.; Giannakouros, T.; Georgatos, S.D. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2001, 2, 920–925.
- Raffa, G.D.; Ciapponi, L.; Cenci, G.; Gatti, M. Terminin: A protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus 2011, 2, 383–391.
- van Bortle, K.; Corces, V.G. Nuclear organization and genome function. Annu. Rev. Cell Dev. Biol. 2012, 28, 163–187.
- Beishline, K.; Vladimirova, O.; Tutton, S.; Wang, Z.; Deng, Z.; Lieberman, P.M. CTCF driven TERRA transcription facilitates completion of telomere DNA replication. Nat. Commun. 2017, 8, 1–10.
- Lis, J.T.; Simon, J.A.; Sutton, C.A. New heat shock puffs and β-galactosidase activity resulting from transformation of Drosophila with an hsp70-lacZ hybrid gene. Cell 1983, 35, 403–410.
- Kellum, R.; Schedl, P. A position-effect assay for boundaries of higher order chromosomal domains. Cell 1991, 64, 941–950.
- Udvardy, A.; Maine, E.; Schedl, P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 1985, 185, 341–358.
- Li, Q.; Starnatoyannopoulos, G. Hypersensitive site 5 of the human B Locus control region functions as a chromatin insulator. Blood 1994, 84, 1399–1401.
- Filippova, G.N.; Fagerlie, S.; Klenova, E.M.; Myers, C.; Dehner, Y.; Goodwin, G.; Neiman, P.E.; Collins, S.J.; Lobanenkov, V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996, 16, 2802–2813.
- Felsenfeld, G.; Bell, A.C. Methylation of a CTCF-dependent boundary controls imprinted expressionof the Igf2 gene. Nature 2000, 405, 482–485.
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680.
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380.
- Brackley, C.A.; Johnson, J.; Michieletto, D.; Morozov, A.; Nicodemi, M.; Cook, P.; Marenduzzo, D. Nonequilibrium chromosome looping via molecular slip links. Phys. Rev. Lett. 2017, 119, 1–5.
- Maksimenko, O.; Bartkuhn, M.; Stakhov, V.; Herold, M.; Zolotarev, N.; Jox, T.; Buxa, M.K.; Kirsch, R.; Bonchuk, A.; Fedotova, A.; et al. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015, 25, 89–99.
- Van Bortle, K.; Nichols, M.H.; Li, L.; Ong, C.T.; Takenaka, N.; Qin, Z.S.; Corces, V.G. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014, 15, R82.
- Yang, J.; Ramos, E.; Corces, V.G. The BEAF-32 insulator coordinates genome organization and function during the evolution of Drosophila species. Genome Res. 2012, 22, 2199–2207.
- Roseman, R.R.; Pirrotta, V.; Geyer, P.K. The su(Hw) protein insulates expression of the drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993, 12, 435–442.
- MacDonald, W.A.; Menon, D.; Bartlett, N.J.; Sperry, G.E.; Rasheva, V.; Meller, V.; Lloyd, V.K. The drosophila homolog of the mammalian imprint regulator, CTCF, maintains the maternal genomic imprint in drosophila melanogaster. BMC Biol. 2010, 8, 105.
- Ohtsuki, S.; Levine, M. GAGA mediates the enhancer blocking activity of the eve promoter in the drosophila embryo. Genes Dev. 1998, 12, 3325–3330.
- Gaszner, M.; Vazquez, J.; Schedl, P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999, 13, 2098–2107.
- Aoki, T.; Sarkeshik, A.; Yates, J.; Schedl, P. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. eLife 2012, 1, 1–24.
- Bag, I.; Dale, R.K.; Palmer, C.; Lei, E.P. The zinc-finger protein CLAMP promotes gypsy chromatin insulator function in Drosophila. J. Cell Sci. 2019, 132, 6.
- Cuartero, S.; Fresán, U.; Reina, O.; Planet, E.; Espinàs, M.L. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 2014, 33, 637–647.
- Gerasimova, T.I.; Gdula, D.A.; Gerasimov, D.V.; Simonova, O.; Corces, V.G. A drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 1995, 82, 587–597.
- Savitsky, M.; Kim, M.; Kravchuk, O.; Schwartz, Y.B. Distinct roles of chromatin insulator proteins in control of the Drosophila bithorax complex. Genetics 2016, 202, 601–617.
- Bartkuhn, M.; Straub, T.; Herold, M.; Herrmann, M.; Rathke, C.; Saumweber, H.; Gilfillan, G.D.; Becker, P.B.; Renkawitz, R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009, 28, 877–888.
- Pherson, M.; Misulovin, Z.; Gause, M.; Dorsett, D. Cohesin occupancy and composition at enhancers and promoters are linked to DNA replication origin proximity in Drosophila. Genome Res. 2019, 29, 602–612.
- Parelho, V.; Hadjur, S.; Spivakov, M.; Leleu, M.; Sauer, S.; Gregson, H.C.; Jarmuz, A.; Canzonetta, C.; Webster, Z.; Nesterova, T.; et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 2008, 132, 422–433.
- Rubio, E.D.; Reiss, D.J.; Welcsh, P.L.; Disteche, C.M.; Filippova, G.N.; Baliga, N.S.; Aebersold, R.; Ranish, J.A.; Krumm, A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. USA. 2008, 105, 8309–8314.
- Holohan, E.E.; Kwong, C.; Adryan, B.; Bartkuhn, M.; Herold, M.; Renkawitz, R.; Russell, S.; White, R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007, 3, 1211–1222.
- Misulovin, Z.; Schwartz, Y.B.; Li, X.Y.; Kahn, T.G.; Gause, M.; MacArthur, S.; Fay, J.C.; Eisen, M.B.; Pirrotta, V.; Biggin, M.D.; et al. Association of cohesin and Nipped-B with transcriptionally active regions of the drosophila melanogaster genome. Chromosoma 2008, 117, 89–102.
- Karch, F.; Galloni, M.; Sipos, L.; Gausz, J.; Gyurkovics, H.; Sched, P. Mcp and Fab-7: Molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of drosophila melanogaster. Nucleic Acids Res. 1994, 22, 3138–3146.
- Cavalli, G.; Paro, R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 1998, 93, 505–518.
- Pérez-Lluch, S.; Cuartero, S.; Azorín, F.; Espinàs, M.L. Characterization of new regulatory elements within the drosophila bithorax complex. Nucleic Acids Res. 2008, 36, 6926–6933.
- Moon, H.; Filippova, G.; Loukinov, D.; Pugacheva, E.; Chen, Q.; Smith, S.T.; Munhall, A.; Grewe, B.; Bartkuhn, M.; Arnold, R.; et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005, 6, 165–170.
- Mohan, M.; Bartkuhn, M.; Herold, M.; Philippen, A.; Heinl, N.; Bardenhagen, I.; Leers, J.; White, A.R.; Renkawitz-Pohl, R.; Saumweber, H.; et al. The drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007, 26, 4203–4214.
- Muller, M.; Hagstrom, K.; Gyurkovics, H.; Pirrotta, V.; Schedl, P. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 1999, 153, 1333–1356.
- Sipos, L.; Gyurkovics, H. Long-distance interactions between enhancers and promoters: The case of the Abd-B domain of the Drosophila bithorax complex. FEBS J. 2005, 272, 3253–3259.
- Kyrchanova, O.; Toshchakov, S.; Podstreshnaya, Y.; Parshikov, A.; Georgiev, P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the drosophila Abd-B gene. Mol. Cell. Biol. 2008, 28, 4188–4195.
- Postika, N.; Metzler, M.; Affolter, M.; Müller, M.; Schedl, P.; Georgiev, P.; Kyrchanova, O. Boundaries mediate long-distance interactions between enhancers and promoters in the drosophila bithorax complex. PLoS Genet. 2018, 14, 1–21.
- Heger, P.; Marin, B.; Bartkuhn, M.; Schierenberg, E.; Wiehe, T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc. Natl. Acad. Sci. USA 2012, 109, 17507–17512.
- Heger, P.; George, R.; Wiehe, T. Successive gain of insulator proteins in arthropod evolution. Evolution (N. Y.) 2013, 67, 2945–2956.
- Seller, C.A.; Cho, C.-Y.; O’Farrell, P.H. Rapid embryonic cell cycles defer the establishment of heterochromatin by Eggless/SetDB1 in drosophila. Genes Dev. 2019, 33, 403–417.
- Armstrong, R.L.; Duronio, R.J. Phasing in heterochromatin during development. Genes Dev. 2019, 33, 379–381.
- Fuks, F.; Hurd, P.J.; Deplus, R.; Kouzarides, T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003, 31, 2305–2312.
- Muramatsu, D.; Kimura, H.; Kotoshiba, K.; Tachibana, M.; Shinkai, Y. Pericentric H3K9me3 Formation by HP1 Interaction-defective Histone Methyltransferase Suv39h1. Cell Struct. Funct. 2016, 41, 145–152.
- Eissenberg, J.C.; Elgin, S.C. The HP1 protein family: Getting a grip on chromation. Curr. Opin. Genet. Dev. 2000, 10, 204.
- Eissenberg, J.C.; Elgin, S.C.R. HP1a: A structural chromosomal protein regulating transcription. Trends Genet. 2014, 30, 103–110.
- Hwang, K.K.; Eissenberg, J.C.; Worman, H.J. Transcriptional repression of euchromatic genes by Drosophila heterochromatin protein 1 and histone modifiers. Proc. Natl. Acad. Sci. USA 2001, 98, 11423–11427.
- Singh, P.B.; Georgatos, S.D. HP1: Facts, open questions, and speculation. J. Struct. Biol. 2002, 140, 10–16.
- Reuter, G.; Giarre, M.; Farah, J.; Gausz, J.; Spierer, A.; Spierer, P. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature 1990, 6263, 219–223.
- Tschiersch, B.; Hofmann, A.; Krauss, V.; Dorn, R.; Korge, G.; Reuter, G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994, 13, 3822–3831.
- Hoskins, R.A.; Smith, C.D.; Carlson, J.W.; Carvalho, A.B.; Halpern, A.; Kaminker, J.S.; Kennedy, C.; Mungall, C.; Sullivan, B.A.; Sutton, G.; et al. Heterochromatic sequences in a drosophila whole-genome shotgun assembly. Genome Biol. 2002, 3, 1–16.
- Hoskins, R.A.; Carlson, J.W.; Kennedy, C.; Acevedo, D.; Evans-Holm, M.; Frise, E.; Wan, K.H.; Park, S.; Mendez-Lago, M.; Rossi, F.; et al. Sequence finishing and mapping of drosophila melanogaster heterochromatin. Science 2007, 316, 1625–1628.
- Ciavatta, D.; Rogers, S.; Magnuson, T. Drosophila CTCF Is Required for Fab-8 enhancer blocking activity in S2 Cells. J. Mol. Biol. 2007, 373, 233–239.
- Bonchuk, A.; Maksimenko, O.; Kyrchanova, O.; Ivlieva, T.; Mogila, V.; Deshpande, G.; Wolle, D.; Schedl, P.; Georgiev, P. Functional role of dimerization and CP190 interacting domains of CTCF protein in drosophila melanogaster. BMC Biol. 2015, 13, 1–23.
- Brower-Toland, B.; Riddle, N.C.; Jiang, H.; Huisinga, K.L.; Elgin, S.C.R. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of drosophila melanogaster. Genetics 2009, 181, 1303–1319.
- Meyer-Nava, S.; Torres, A.; Zurita, M.; Valadez-Graham, V. Molecular effects of dADD1 misexpression in chromatin organization and transcription. BMC Mol. Cell Biol. 2020, 2, 1–17.
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245.
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, 460–464.
- Frydrychova, R.C.; Biessmann, H.; Mason, J.M. Regulation of telomere length in Drosophila. Cytogenet. Genome Res. 2009, 122, 356–364.
- Gibbons, R.J.; Picketts, D.J.; Villard, L.; Higgs, D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 1995, 80, 837–845.
- Sadic, D.; Schmidt, K.; Groh, S.; Kondofersky, I.; Ellwart, J.; Fuchs, C.; Theis, F.J.; Schotta, G. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 2015, 16, 836–850.
- Valle-García, D.; Qadeer, Z.A.; McHugh, D.S.; Ghiraldini, F.G.; Chowdhury, A.H.; Hasson, D.; Dyer, M.A.; Recillas-Targa, F.; Bernstein, E. ATRX binds to atypical chromatin domains at the 3′ exons of zinc finger genes to preserve H3K9me3 enrichment. Epigenetics 2016, 11, 398–414.
- de Wit, E.; Greil, F.; van Steensel, B. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 2007, 3, 0346–0357.
- Ilyin, A.A.; Stolyarenko, A.D.; Klenov, M.S.; Shevelyov, Y.Y. Various modes of HP1a interactions with the euchromatic chromosome arms in Drosophila ovarian somatic cells. Chromosoma 2020, doi:10.1007/s00412-020-00738-5.
- Li, Y.; Danzer, J.R.; Alvarez, P.; Belmont, A.S.; Wallrath, L.L. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 2003, 130, 1817–1824.
- Piacentini, L.; Fanti, L.; Berloco, M.; Perrini, B.; Pimpinelli, S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 2003, 161, 707–714.
- Petesch, S.J.; Lis, J.T. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 Loci. Cell 2008, 134, 74–84.
- Schneiderman, I.J.; Orsi, G.A.; Hughes, K.T.; Loppin, B.; Ahmad, K. Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc. Natl. Acad. Sci. USA 2012, 109, 19721–19726.
- Lu, B.Y.; Emtage, P.C.; Duyf, B.J.; Hilliker, A.J.; Eissenberg, J.C. heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in drosophila. Genetics 2000, 155, 699–708.
- Danzer, J.R.; Wallrath, L.L. Mechanisms of HP1-mediated gene silencing in Drosophila. Development 2004, 131, 3571–3580.
- de Lucia, F.; Ni, J.Q.; Vaillant, C.; Sun, F.L. HP1 modulates the transcription of cell-cycle regulators in drosophila melanogaster. Nucleic Acids Res. 2005, 33, 2852–2858.
- Minc, E.; Courvalin, J.; Buendia, B. HP1Ɣ associates associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2005, 284, 279–284.
- Vakoc, C.R.; Mandat, S.A.; Olenchock, B.A.; Blobel, G.A. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 2005, 19, 381–391.
- Kwon, S.H.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Abmayr, S.M.; Workman, J.L. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev. 2010, 24, 2133–2145.
- Lee. Y.C.G.; Ogiyama, Y.; Martins, N.M.C; Beliveau, B.J.; Acevedo, D.; Wu, C-T.Cavalli, G.; Karpen, G.H.; et al. Pericentromeric heterochromatin is hierarchically organized and spatially contacts H3K9me2 islands in euchromatin. PLoS Genet. 2020, 16, 1–27.
