Leptospirosis is a globally distributed zoonotic disease caused by pathogenic bacteria of the genus Leptospira. This zoonotic disease affects humans, domestic animals, and wild animals. Colombia is considered an endemic country for leptospirosis, and Antioquia is the second department in Colombia, with the highest number of reported leptospirosis cases.
- Leptospira
- bats
- Colombia
- leptospirosis
- species
- type
1. Introduction
Leptospirosis is an emerging zoonotic disease caused by pathogenic bacteria of the genus Leptospira [1]. Previous studies have estimated that 1.03 million cases and 58,900 deaths occur due to leptospirosis worldwide annually [2]. Leptospirosis is considered a neglected disease, found mainly in the tropical regions of developing countries [3] and is now recognized as an emerging infectious disease due to large outbreaks in different regions of the world, which are associated with environmental disasters, and extreme climate change. In many endemic regions, severe forms of the disease, such as Weil’s disease and pulmonary hemorrhage syndrome have emerged as the leading cause of death [4]. Currently, about 65 genomic Leptospira species have been identified (NCBI database: https://www.ncbi.nlm.nih.gov/genome [accessed on 30 April 2021]), which are subdivided into four main clades according to the phylogenetic analysis of 1371 conserved genes: pathogens (P1), pathogens (P2), saprophytes (S1), and saprophytes (S2) [4][5]. Through serological classification about 300 Leptospira serovars have been described, which are grouped into approximately 30 serogroups and about 200 of these serovars have been considered pathogenic [6]. Colombia is an endemic country for leptospirosis with at least 500 cases every year [7]. Antioquia is the second department in Colombia with the highest number of confirmed cases of leptospirosis [7], with a seroprevalence close to 12.5% [8]. Leptospira interrogans and L. santarosai have been identified as the causative agents of this disease [9]. Therefore, this department in an important region in Colombia for the study of leptospirosis.
Bats have been identified worldwide as an important reservoir of different Leptospira species (L. interrogans, L. borgpetersenii, L. kirschneri, L. fainei) and their role in disease transmission, and spillover in the life cycle of this bacterium has yet to be defined [22]. Currently, more than 50 species of infected bats with Leptospira have been reported in different countries, including Peru [23], Brazil [24], Argentina [25], Australia [26], Comoros island and Madagascar [27], Reunion Island [28], Mayotte Island [22], Indonesia [23], Malaysia [24], Tanzania [25], Trinidad [26], Sudan [27], Democratic Republic of Congo [28], Africa [29], and Azerbaijan [30]. In Colombia, two studies have reported the presence of bats naturally infected with Leptospira [31][32]. Due to the above characteristics, bats could act as excellent spillover of Leptospira species to the environment, favoring contamination of water and soil, serving as a direct or indirect source of infection for other animals, which are the main reservoirs and disseminators of the bacteria. The objective of the present investigation was to detect Leptospira species infecting different bat species in the Urabá region (Antioquia-Colombia) and to evaluate the genetic diversity of the circulating Leptospira species. This information will illustrate the role of bats in the transmission cycle of human leptospirosis.
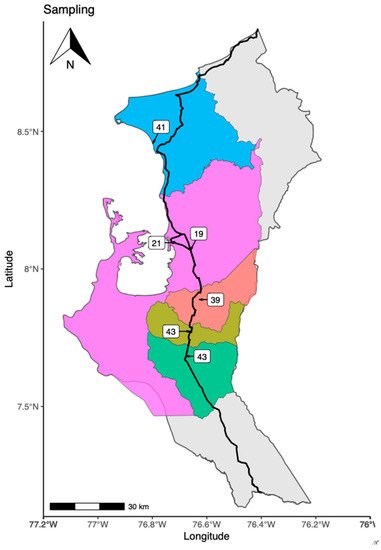
2. Places of Bat’s Capture
3. Families, Genera and Species of Captured Bats
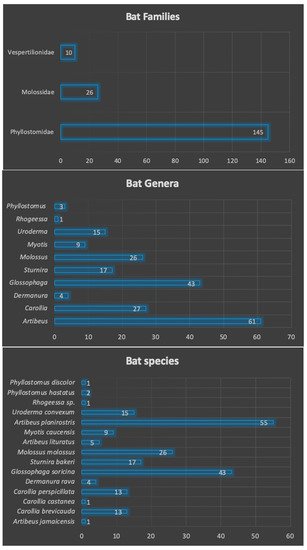
| Species | Number | Percentage (%) | Frequency | Feeding Habits |
|---|---|---|---|---|
| Artibeus jamaicensis | 1 | 0.49% | 0.005 | frugivore |
| Carollia brevicauda | 13 | 6.31% | 0.063 | frugivore |
| Carollia castanea | 1 | 0.49% | 0.005 | frugivore |
| Carollia perspicillata | 13 | 6.31% | 0.063 | frugivore |
| Dermanura rava | 4 | 1.94% | 0.019 | frugivore |
| Glossophaga soricina | 43 | 20.87% | 0.209 | nectarivore |
| Sturnira bakeri | 17 | 8.25% | 0.083 | frugivore |
| Molossus molossus | 26 | 12.62% | 0.126 | insectivore |
| Artibeus lituratus | 5 | 2.43% | 0.024 | frugivore |
| Myotis caucensis | 9 | 4.37% | 0.044 | insectivore |
| Artibeus planirostris | 55 | 26.70% | 0.267 | frugivore |
| Uroderma convexum | 15 | 7.28% | 0.073 | frugivore |
| Rhogeessa sp. | 1 | 0.49% | 0.005 | insectivore |
| Phyllostomus hastatus | 2 | 0.97% | 0.010 | omnivore |
| Phyllostomus discolor | 1 | 0.49% | 0.005 | omnivore |
| TOTAL | 206 | 100% | 1 |
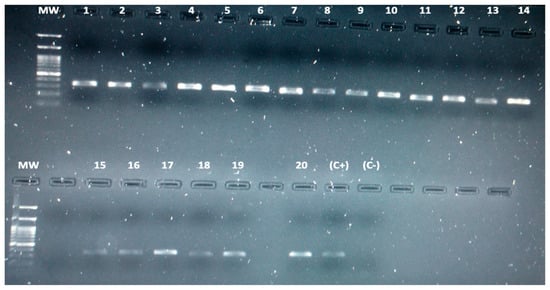
4. Detection of Leptospira spp. in Bats by Conventional PCR
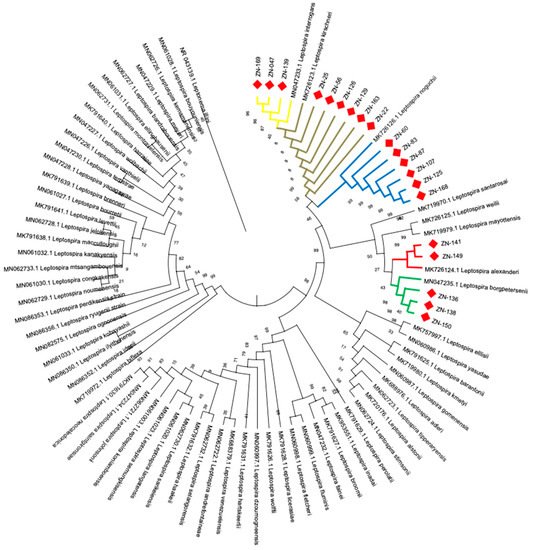
| Code | Phylogenetic Identification (16S Ribosomal Gene) |
Infected Species | Feeding Habits | Gender | Municipality |
|---|---|---|---|---|---|
| ZM-022 | Leptospira kirschneri | Carollia perspicillata | Frugivore | Female | Carepa |
| ZM-025 | Leptospira kirschneri | Dermanura rava | Frugivore | Male | Carepa |
| ZM-047 | Leptospira interrogans | Glossophaga soricina | Nectarivore | Female | Apartadó |
| ZM-056 | Leptospira kirschneri | Glossophaga soricina | Nectarivore | Male | Apartadó |
| ZM-060 | Leptospira noguchii | Glossophaga soricina | Nectarivore | Female | Apartadó |
| ZN-083 | Leptospira noguchii | Uroderma convexum | Frugivore | Male | Chigorodó |
| ZN-087 | Leptospira noguchii | Uroderma convexum | Frugivore | Male | Chigorodó |
| ZN-107 | Leptospira noguchii | Uroderma convexum | Frugivore | Female | Chigorodó |
| ZN-125 | Leptospira noguchii | Molossus molossus | Insectivore | Female | Turbo |
| ZN-126 | Leptospira kirschneri | Molossus molossus | Insectivore | Male | Turbo |
| ZN-129 | Leptospira kirschneri | Artibeus planirostris | Frugivore | Male | Turbo |
| ZN-136 | Leptospira borgpetersenii | Glossophaga soricina | Nectarivore | Female | Turbo |
| ZN-138 | Leptospira borgpetersenii | Glossophaga soricina | Nectarivore | Female | Turbo |
| ZN-139 | Leptospira interrogans | Artibeus planirostris | Frugivore | Male | Turbo |
| ZN-141 | Leptospira alexanderi | Uroderma convexum | Frugivore | Female | Turbo |
| ZN-149 | Leptospira alexanderi | Glossophaga soricina | Nectarivore | Male | Turbo |
| ZN-150 | Leptospira borgpetersenii | Artibeus planirostris | Frugivore | Male | Turbo |
| ZN-163 | Leptospira kirschneri | Artibeus planirostris | Frugivore | Male | Turbo |
| ZN-168 | Leptospira noguchii | Uroderma convexum | Frugivore | Male | Necoclí |
| ZN-169 | Leptospira interrogans | Uroderma convexum | Frugivore | Female | Necoclí |
5. Identification of Leptospira Species by Phylogenetic Analysis
6. Host-Pathogen Relationship between Bats and Leptospira
| Leptospira Species | Infected Bat Species | Infected Bats |
|---|---|---|
| Leptospira borgpetersenii |
Glossophaga soricina (2)Artibeus planirostris (1) |
3 |
| Leptospira alexanderi |
Uroderma convexum (1)Glossophaga soricina (1) |
2 |
| Leptospira noguchii |
Glossophaga soricina (1)Uroderma convexum (4)Molossus molossus (1) |
6 |
| Leptospira interrogans |
Glossophaga soricina (1)Artibeus planirostris (1)Uroderma convexum (1) |
3 |
| Leptospira kirschneri |
Carollia perspicillata (1)Dermanura rava (1)Glossophaga soricina (1)Molossus molossus (1)Artibeus planirostris (2) |
6 |
Total: 20 |
7. Conclusion
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms9091897
References
- Adler, B.; de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296.
- Costa, F.; Hagan, J.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global morbidity and mortality of leptospirosis: A systematic review. PLoS Negl. Trop. Dis. 2015, 9, e0003898.
- Haake, D.A.; Levett, P.N. Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97.
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neel, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Khalid, M.K.N.M.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270.
- Cerqueira, G.M.; Picardeau, M. A century of Leptospira strain typing. Infect. Genet. Evol. 2009, 9, 760–768.
- Bello, S.; Rodríguez, M.; Paredes, A.; Mendivelso, F.; Walteros, D.; Rodríguez, F.; Realpe, M.E. Comportamiento de la vigilancia epidemiológica de la leptospirosis humana en Colombia, 2007–2011. Biomédica 2013, 33 (Suppl. 1), 153–160.
- Carreño, L.A.; Salas, D.; Beltrán, K.B. Prevalencia de leptospirosis en Colombia: Revisión sistemática de literatura. Prevalence of leptospirosis in Colombia: Systematic literature review. Rev. Salud Pública 2017, 19, 204–209.
- Agudelo, F.P.; Restrepo-Jaramillo, B.N.; Arboleda-Naranjo, M. Situación de la leptospirosis en el Urabá antioqueño colombiano: Estudio seroepidemiológico y factores de riesgo en población general urbana. Cad. Saúde Pública 2007, 23, 2094–2102.
- Sanchez, R.G.P.; Lopez, J.; Pereira, M.M.; Naranjo, M.A.; Agudelo-Flórez, P. Genetic diversity of Leptospira in northwestern Colombia: First report of Leptospira santarosai as a recognised leptospirosis agent. Mem. Inst. Oswaldo Cruz. 2016, 111, 737–744.
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545.
- Mühldorfer, K. Bats and bacterial pathogens: A review. Zoonoses Public Health 2013, 60, 93–103.
- Juliane Schaer, J.; Perkins, S.L.; Decher, J.; Leendertz, F.H.; Fahr, J.; Weber, N.; Kai Matuschewski, K. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc. Natl. Acad. Sci. USA 2013, 110, 17415–17419.
- Mok, W.Y.; Luizão, R.C.; Barreto da Silva, M.S. Isolation of fungi from bats of the Amazon basin. Appl. Environ. Microbiol. 1982, 44, 570–575.
- Griffin, D.R. Migrations and homing in bats. In Biology of Bats, 2nd ed.; Wimsatt, W.A., Ed.; Academic Press: New York, NY, USA, 1970; pp. 233–264.
- Amador, L.I.; Moyers-Arévalo, R.L.; Almeida, F.C.; Catalano, S.A.; Giannini, N.P. Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. J. Mammal. Evol. 2018, 25, 37–70.
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’Brien, S.J.; Murphy, W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 2005, 307, 580–584.
- Holland, R.A.; Waters, D.A.; Rayner, J.M. Echolocation signal structure in the megachiropteran bat Rousettus aegyptiacus Geoffroy 1810. J. Exp. Biol. 2004, 207, 4361–4369.
- Lyman, C.P. Thermoregulation and metabolism in bats. In Biology of Bats, 2nd ed.; Wimsatt, W.A., Ed.; Academic Press: New York, NY, USA, 1970; pp. 301–330.
- Constantine, D.G. Activity Patterns of the Mexican Free-Tailed Bat; University of New Mexico Press: Albuquerque, NM, USA, 1967; pp. 1–79.
- Cockrum, E.L. Migration in the Guano Bat, Tadarida Brasiliensis; University of Kansas Museum of Natural History: Lawrence, KS, USA, 1969; pp. 303–336.
- Eisenberg, J.F. The density and biomass of tropical mammals. In Conservation Biology; Soule, M., Wilcox, B.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1980; pp. 35–56.
- Desvars, A.; Naze, F.; Vourc’h, G.; Cardinale, E.; Picardeau, M.; Bourhy, P. Similarities in Leptospira serogroup and species distribution in animals and humans in the Indian ocean island of Mayotte. Am. J. Trop. Med. Hyg. 2021, 87, 134–140.
- Van Peenen, P.F.D.; Light, R.H.; Sulianti-Saroso, J. Leptospirosis in wild mammals of Indonesia-Recent surveys. Southeast Asian J. Trop. Med. Public Health 1971, 2, 496–502.
- Thayaparan, S.; Robertson, I.A.N.; Amraan, F.; Ut, L.S.U.; Abdullah, M.T. Serological prevalence of leptospiral Infection in wildlife in Sarawak, Malaysia. Borneo J. Res. Sci. Technol. 2013, 2, 71–74.
- Mgode, G.F.; Mbugi, H.A.; Mhamphi, G.G.; Ndanga, D.; Nkwama, E.L. Seroprevalence of Leptospira infection in bats roosting in human settlements in Morogoro municipality in Tanzania. Tanzan. J. Health Res. 2014, 16, 1–7.
- Everard, C.R.; Fraser-Chanpong, G.M.; Bhagwandin, L.J.; Race, M.W.; James, A.C. Leptospires in wildlife from Trinidad and Grenada. J. Wildl. Dis. 1983, 19, 192–199.
- Sebek, Z.; Sixl, W.; Reinthaler, F.; Valova, M.; Scneeweiss, W.; Stünzner, D.; Mascher, F. Results of serological examination for leptospirosis of domestic and wild animals in the Upper Nile province (Sudan). J. Hyg. Epidemiol. Microb. Immunol. 1989, 33, 337–345.
- Ogawa, H.; Koizumi, N.; Ohnuma, A.; Mutemwa, A.; Hang, B.M.; Mweene, A.S.; Takada, A.; Sugimoto, C.; Suzuki, Y.; Kida, H.; et al. Molecular epidemiology of pathogenic Leptospira spp. in the straw-colored fruit bat (Eidolon helvum) migrating to Zambia from the Democratic Republic of Congo. Infect. Genet. Evol. 2015, 32, 143–147.
- Jobbins, S.E.; Alexander, K.A. Evidence of Leptospira sp. infection among a diversity of African wildlife species: Beyond the usual suspects. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 349–351.
- Tagi-Zade, T.A.; Mardanly, A.S.; Akhmedov, I.B.; Alekperov, F.P.; Gasanov, S.N. Examination of bats for leptospirosis in the territory of Azerbaijan SSR. Zhurnal Mikrobiol. Epidemiol. I Immunobiol. 1970, 9, 118–121.
- Victoria, R.J.; Iriarte, L.J.; Sampedro, A.C. Presence of Leptospira spp. in urban bats from Sincelejo, Sucre, Colombia. Int. J. Pharm. Tech. Res. 2018, 11, 218–225.
- Mateus, J.; Gómez, N.; Herrera-Sepúlveda, M.T.; Hidalgo, M.; Pérez-Torres, J.; Cuervo, C. Bats are a potential reservoir of pathogenic Leptospira species in Colombia. J. Infect. Dev. Ctries. 2019, 13, 278–283.
