Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Mitochondria are subcellular organelles involved in essential cellular functions, including cytosolic calcium regulation, cell apoptosis, and reactive oxygen species production. They are the site of important biochemical pathways, including the tricarboxylic acid cycle, parts of the ureagenesis cycle, or haem synthesis.
- mitochondria
- ROS
- diabetes
- obesity
- cardiovascular disease
- antioxidants
1. The Electron Transport Chain
The mitochondrion is a cytoplasmic organelle that consists of a double membrane, matrix, and mtDNA. The outer membrane and intermembrane space are relatively permeable in contrast to the inner membrane which has a restrictive permeability, containing the enzymes required for electron transport [1]. Mitochondria generate the majority of cellular energy in the form of ATP, through the oxidation of reduced nicotinamide adenine dinucleotide (NADH) and reduced flavin adenine dinucleotide (FADH2), and subsequently, the process of oxidative phosphorylation [2]. Molecules derived from the catabolism of glucose (glycolysis), fatty acids (beta-oxidation), and amino acids (deamination/transamination) are further referred to the tricarboxylic acid (TCA) cycle to generate the required OXPHOS substrate [3]. An electrochemical gradient generated at the level of the inner membrane generates OXPHOS. The electron transport chain (ETC) is made up of five enzyme complexes (I, II, III, IV, and V), located at the level of the mitochondrial inner membrane. Electrons donated by NADH/FADH2 coenzymes are transferred to complex I (NADH: ubiquinone reductase) or complex II (succinate dehydrogenase) of the ETC [4]. The two electrons from NADH are given to ubiquinone (UQ) with the help of cofactors. Subsequently, ubiquinone is reduced to ubiquinol (UQH2). This transfer of electrons triggers the introduction of protons from the matrix into the intermembrane space (through the transfer of two electrons, four protons are introduced). Electrons donated by FADH2 are transferred to the UQ via complex II but are not associated with the transport of protons from the matrix into the intermembrane space [3]. Afterward, they are transferred to complex III (cytochrome c reductase), made up of cytochromes b and c1. The entire process of electron transfer from UQH2 to cytochrome c is called the Q cycle. Initially, UQH2 binds to complex III, facilitating the access of two protons in the intermembrane space, while two electrons are released, following different paths. The first electron is transferred to cytochrome c1 (at this level it reduces Fe3+ to Fe2+), and from this level, it is then transferred to cytochrome c. The second electron is given to cytochrome b; subsequently, UQ is partially reduced to a molecule called the semi-quinone radical ion (Q-). In the second stage, a new UQH2 molecule is attached to complex III following the same pattern; therefore, a new electron is bound to the cytochrome c level, and the second electron is bound at the Q level with the formation of a UQH2 molecule. At the end of this process, four protons are generated in the intermembrane space. Four electrons are transferred from four cytochrome c molecules to complex IV (cytochrome c oxidase), where molecular oxygen is bound and reduced to water. Finally, at the level of complex IV, eight protons are transferred from the matrix (four are used for the formation of two water molecules, and the other four are transferred to the intermembrane space) [5]. At the end of the electron transport process, using one molecule of NADH, 10 protons are generated towards the intermembrane space (two from complex IV and four each from complex I and complex III, respectively). In this way, an electrochemical gradient known as mitochondrial membrane potential is produced. Complex V (F0F1 ATP synthase) consists of two domains: extramembrane (F1) and transmembrane (F0). This transport of electrons is associated with the transport of protons from the level of the internal membrane, generating the electrochemical gradient that is necessary for ATP production [3][4][6] (Figure 1).
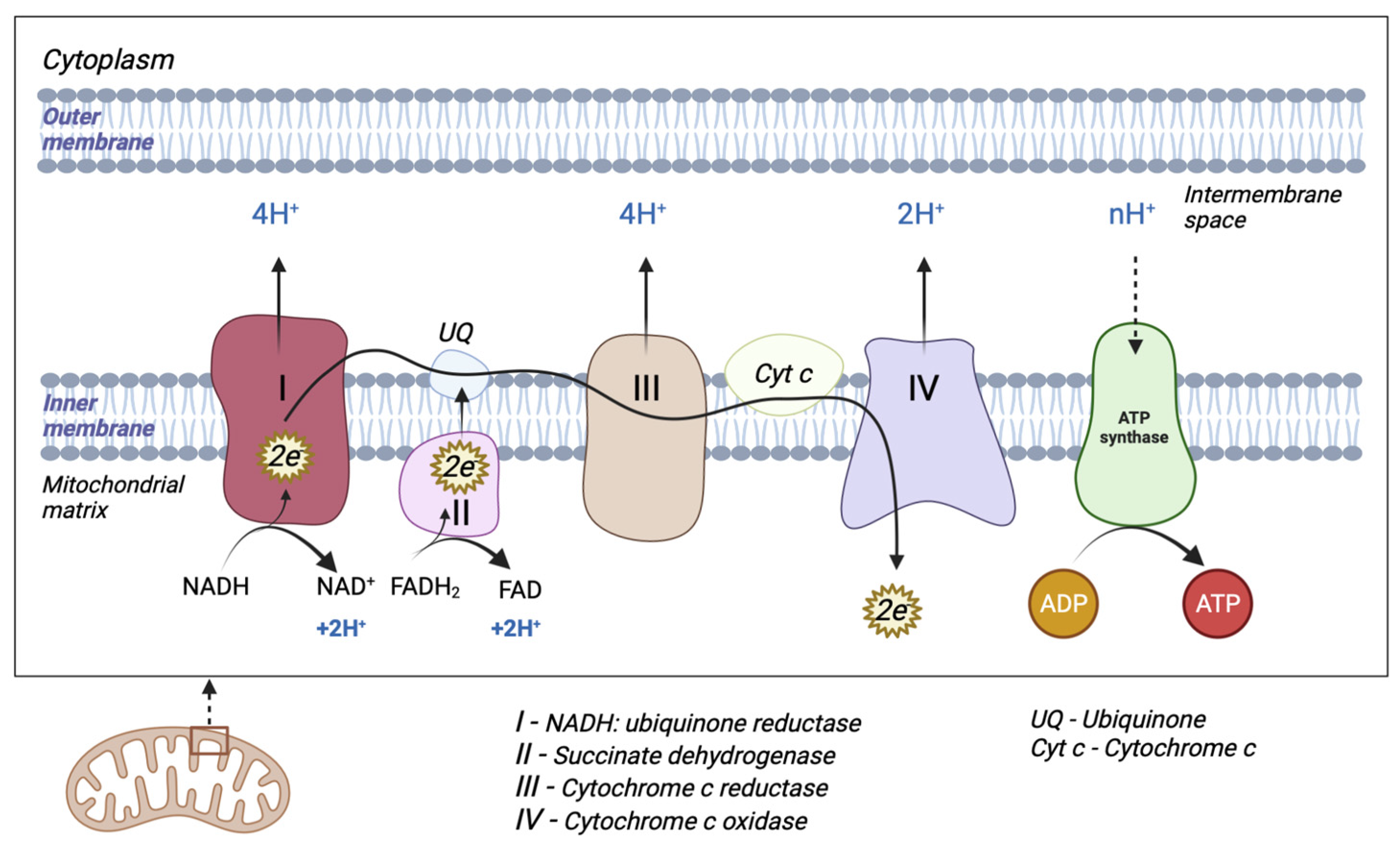
Figure 1. Schematic representation of mitochondrial electron transport chain (ETC). The ETC consists of five enzyme complexes (I, II, III, IV, and V).
2. Mitochondrial DNA Structure
mtDNA nucleotide sequences were first identified in 1981, and were further re-evaluated and subsequently revised in 1999 [7][8]. mtDNA is a double-stranded circular DNA molecule consisting of 16,569 bp which encodes 37 genes, including 13 polypeptides essential for the OXPHOS mechanism, 2 ribosomal RNAs (12S and 16S), and 22 transfer RNAs. mtDNA has a special structure compared to genomic DNA; it does not contain introns, as genes have absent or reduced portions of non-coding bases between them [9].
Zong et al. described free circulating mtDNA in blood samples with an important prognostic role in various cancers, cardiac arrest, and sepsis [10]. Subsequently, circulating free mtDNA was identified as a major mediator of innate immunity and systemic inflammatory response. The process of being released into plasma results in the activation of neutrophils, mediated by the Toll-like receptor 9 (TLR9) [11]. mtDNA is also found in the cytosol. It has been shown that oxidative stress, viral or bacterial infections, or miss-packaging lead to its release and are involved in innate intracellular immune responses [12]. Mitochondrial dysfunctions have been correlated with obesity, diabetes mellitus, and cardiovascular pathologies. An increased amount of glucose is predisposed to the increased production of ROS, with destructive effects at the mitochondrial level [13]. The aging process, the reduced action capacity of antioxidants, and the changes produced at the mitochondrial level can be as important causes of metabolic pathologies.
3. Mitochondrial Biogenesis and Dynamics
Most mitochondrial proteins are nuclear-encoded proteins and are translated by cytosolic ribosomes, processed, and imported into the mitochondria via the TIM/TOM system [14]. The TOM complex is the translocase of the outer mitochondrial membrane and mediates the importing of nuclear-encoded proteins into the intermembrane space [15]. There are two distinct mitochondrial translocase complexes in the inner mitochondrial membrane (TIM) [16]. The TIM22 and TIM23 complexes recognize and import different classes of proteins [17]. Mitochondrial dynamics is essential in maintaining mitochondrial homeostasis and is achieved through two processes: fusion and fission. Imbalances between the two events generate mitochondrial morphological changes, an excess of fission causes the formation of fragmented mitochondria, and an excess of fusion triggers mitochondria elongation.
Mitofusins (Mfn) 1 and 2 are proteins involved in the fusion process of the outer mitochondrial membrane. The fusion of the outer mitochondrial membrane is most often achieved simultaneously with the fusion of the inner membrane, with the latter being mediated by the optic atrophy 1 protein (OPA1). The absence of Mfn cuts off the fusion phenomenon of both membranes. Mitochondrial fission is regulated by dynamin-related protein 1 (Drp1) and fission protein (Fis1) [18]. Under various metabolic conditions, several disbalances in such proteins occur during hyperglycaemic conditions, and Drp1 and Fis1 are increased, while Mfn1, Mfn2, and OPA1 are reduced [19].
Mitochondrial biogenesis is a complex process through which cells increase their mitochondrial mass and require coordination between nuclear and mitochondrial DNA. This process involves mtDNA transcription and translation processes, and the synthesis, import, and association of mitochondrial proteins encoded by nuclear DNA [20].
Mitochondrial biogenesis dysfunction has been associated with metabolic disorders such as obesity and T2DM. A decline in the proliferator-activated receptor gamma coactivator-1α (PGC-1α), AMP-activated protein kinase (AMPK), and silent information regulator 1 (SIRT-1) signalling pathways seems to be the underlying mechanism for reduced mitochondrial biogenesis in the diabetic kidney and the diabetic heart as well, with hypoadiponectinemia being reported to impair AMPK-PGC-1α signalling [20].
4. Mitophagy
Autophagy is a natural mechanism which was highly conservated throughout evolution, by which the useless cytoplasmic material is transported to lysosomes for destruction [21]. Autophagy is influenced by a variety of factors. The autophagic response promotes the adaptation to stress and increases cellular viability [22]. Components of the autophagy response are implicated in regulated cell death [23].
The degradation of mitochondria through selective autophagy is referred to as mitophagy, a process that involves the selective sequestration of damaged or dysfunctional mitochondria into double-membraned autophagosomes for later lysosomal destruction. Mitophagy has been described in mammalian cells as being facilitated by two well-studied pathways, ubiquitin-mediated and receptor-mediated, and is essential for maintaining cellular fitness [24][25].
Mitophagy ubiquitin-mediated pathways are regulated by two key proteins PTEN-induced putative kinase protein 1 (PINK1) and Parkin. Normally, PINK1 is imported into healthy mitochondria via the TIM/TOM system and further degraded by proteolytic reactions. Damaged mitochondria lose membrane potential, which impairs the TIM/TOM system’s function, resulting in the accumulation of PINK1 on the outer mitochondrial membrane, which promotes the recruitment of Parkin and the activation of its ubiquitination ligase activity, leading to the ubiquitination of proteins from the outer mitochondrial membrane. Further, Parkin promotes the recruitment of autophagy adaptors, such as optineurin (OPTN) and nuclear dot protein 52 kDa (NDP52), leading to the degradation of damaged mitochondria [26][27][28].
The mitophagy receptor pathway is mediated by receptors embedded in the outer mitochondrial membrane, most notably by NIX (known as BCL2 interacting protein 3 like (BNIP3L)), BCL2 interacting protein 3 (BNIP3), and FUN14 domain containing 1 (FUNDC1), which are characterized by the presence of an LC3-interacting region (LIR) that can directly bind to the autophagy mediator LC3 to promote mitophagy when mitochondria are damaged [24][29].
Mitophagy is implicated in insulin resistance and some cardiac pathological conditions. The dysfunctional mitophagy mechanism has been linked to the development of insulin resistance [30]. Moreover, an efficient mitophagical response helps the cardiomyocytes to survive during the nutritional stress in myocardial infarction [31].
This entry is adapted from the peer-reviewed paper 10.3390/antiox12030658
References
- Sherratt, H.S. Mitochondria: Structure and Function. Rev. Neurol. 1991, 147, 417–430.
- Chen, X.J.; Butow, R.A. The Organization and Inheritance of the Mitochondrial Genome. Nat. Rev. Genet. 2005, 6, 815–825.
- Zhao, R.-Z.; Jiang, S.; Zhang, L.; Yu, Z.-B. Mitochondrial Electron Transport Chain, ROS Generation and Uncoupling (Review). Int. J. Mol. Med. 2019, 44, 3–15.
- Dallner, G.; Sindelar, P.J. Regulation of Ubiquinone Metabolism. Free Radic. Biol. Med. 2000, 29, 285–294.
- Ahmad, M.; Wolberg, A.; Kahwaji, C.I. Biochemistry, Electron Transport Chain. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022.
- Andreyev, A.Y.; Kushnareva, Y.E.; Starkov, A.A. Mitochondrial Metabolism of Reactive Oxygen Species. Biochemistry 2005, 70, 200–214.
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and Organization of the Human Mitochondrial Genome. Nature 1981, 290, 457–465.
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and Revision of the Cambridge Reference Sequence for Human Mitochondrial DNA. Nat. Genet. 1999, 23, 147.
- Protasoni, M.; Zeviani, M. Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions. Int. J. Mol. Sci. 2021, 22, 586.
- Zong, W.-X.; Rabinowitz, J.D.; White, E. Mitochondria and Cancer. Mol. Cell 2016, 61, 667–676.
- Shokolenko, I.; Alexeyev, M. Mitochondrial DNA: Consensuses and Controversies. DNA 2022, 2, 131–148.
- West, A.P.; Khoury-Hanold, W.; Staron, M.; Tal, M.C.; Pineda, C.M.; Lang, S.M.; Bestwick, M.; Duguay, B.A.; Raimundo, N.; MacDuff, D.A.; et al. Mitochondrial DNA Stress Primes the Antiviral Innate Immune Response. Nature 2015, 520, 553–557.
- Kowaltowski, A.J.; de Souza-Pinto, N.C.; Castilho, R.F.; Vercesi, A.E. Mitochondria and Reactive Oxygen Species. Free Radic. Biol. Med. 2009, 47, 333–343.
- Yapa, N.M.B.; Lisnyak, V.; Reljic, B.; Ryan, M.T. Mitochondrial Dynamics in Health and Disease. FEBS Lett. 2021, 595, 1184–1204.
- Araiso, Y.; Imai, K.; Endo, T. Role of the TOM Complex in Protein Import into Mitochondria: Structural Views. Annu. Rev. Biochem. 2022, 91, 679–703.
- Tang, K.; Zhao, Y.; Li, H.; Zhu, M.; Li, W.; Liu, W.; Zhu, G.; Xu, D.; Peng, W.; Xu, Y.-W. Translocase of Inner Membrane 50 Functions as a Novel Protective Regulator of Pathological Cardiac Hypertrophy. J. Am. Heart Assoc. 2017, 6, e004346.
- Bauer, M.F.; Hofmann, S.; Neupert, W.; Brunner, M. Protein Translocation into Mitochondria: The Role of TIM Complexes. Trends Cell Biol. 2000, 10, 25–31.
- van der Bliek, A.M.; Shen, Q.; Kawajiri, S. Mechanisms of Mitochondrial Fission and Fusion. Cold Spring Harb. Perspect. Biol. 2013, 5, a011072.
- Kaikini, A.A.; Kanchan, D.M.; Nerurkar, U.N.; Sathaye, S. Targeting Mitochondrial Dysfunction for the Treatment of Diabetic Complications: Pharmacological Interventions through Natural Products. Pharmacogn. Rev. 2017, 11, 128–135.
- Popov, L.-D. Mitochondrial Biogenesis: An Update. J. Cell. Mol. Med. 2020, 24, 4892–4899.
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular Definitions of Autophagy and Related Processes. EMBO J. 2017, 36, 1811–1836.
- Fuchs, Y.; Steller, H. Live to Die Another Way: Modes of Programmed Cell Death and the Signals Emanating from Dying Cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 329–344.
- Mariño, G.; Niso-Santano, M.; Baehrecke, E.H.; Kroemer, G. Self-Consumption: The Interplay of Autophagy and Apoptosis. Nat. Rev. Mol. Cell Biol. 2014, 15, 81–94.
- Guan, Y.; Wang, Y.; Li, B.; Shen, K.; Li, Q.; Ni, Y.; Huang, L. Mitophagy in Carcinogenesis, Drug Resistance and Anticancer Therapeutics. Cancer Cell Int. 2021, 21, 350.
- Ma, K.; Chen, G.; Li, W.; Kepp, O.; Zhu, Y.; Chen, Q. Mitophagy, Mitochondrial Homeostasis, and Cell Fate. Front. Cell Dev. Biol. 2020, 8, 467.
- Iorio, R.; Celenza, G.; Petricca, S. Mitophagy: Molecular Mechanisms, New Concepts on Parkin Activation and the Emerging Role of AMPK/ULK1 Axis. Cells 2021, 11, 30.
- Kim, S.-J.; Ahn, D.-G.; Syed, G.H.; Siddiqui, A. The Essential Role of Mitochondrial Dynamics in Antiviral Immunity. Mitochondrion 2018, 41, 21–27.
- Mitophagy Regulated by the PINK1-Parkin Pathway|IntechOpen. Available online: https://www.intechopen.com/chapters/49196 (accessed on 26 January 2023).
- Killackey, S.A.; Philpott, D.J.; Girardin, S.E. Mitophagy Pathways in Health and Disease. J. Cell Biol. 2020, 219, e202004029.
- Shan, Z.; Fa, W.H.; Tian, C.R.; Yuan, C.S.; Jie, N. Mitophagy and Mitochondrial Dynamics in Type 2 Diabetes Mellitus Treatment. Aging 2022, 14, 2902–2919.
- Riquelme, J.A.; Chavez, M.N.; Mondaca-Ruff, D.; Bustamante, M.; Vicencio, J.M.; Quest, A.F.G.; Lavandero, S. Therapeutic Targeting of Autophagy in Myocardial Infarction and Heart Failure. Expert Rev. Cardiovasc. Ther. 2016, 14, 1007–1019.
This entry is offline, you can click here to edit this entry!
