Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biotechnology & Applied Microbiology
Chagas disease, caused by Trypanosoma cruzi infections, is included in the group of neglected diseases, and efforts to develop new therapeutic or immunoprevention approaches have not been successful. After the publication of the T. cruzi genome, the number of molecular and biochemical studies on this parasite has increased considerably, many of which are focused on families of variant surface proteins, especially trans-sialidases, mucins, and mucin-associated proteins. The disperse gene protein 1 family (DGF-1) is one of the most abundant families in the T. cruzi genome.
- chagas disease
- surface proteins
- disperse protein family
- Trypanosoma cruzi
1. Introduction
Trypanosoma cruzi, the etiological agent of American Trypanosomiasis or Chagas disease, has proven to be a big challenge for those working on the molecular biology and genomics of the parasite. The repetitive nature of the T. cruzi genome, the presence of cell hybridization events, variations in ploidy, and chromosomal size polymorphisms have led to heated debates about the nature of its population dynamics [1]. Nonetheless, T. cruzi is a pathogenic entity, and depending on geographic locations and routes of infection, the symptoms in the human host vary [2]. Chagas is currently reported from the south of the United States to Argentina, and due to human migration, cases are also registered in non-endemic regions of Europe and Asia [3]. Attempts to develop new effective drugs or vaccines have failed, and the only drugs for treatment are nifurtimox and benznidazole. The parasite’s resilience and variability rely on its very dynamic genome, making it necessary to search for new ways to control this parasite [4]. The Disperse Gene Family (DGF-1) ranked fifth among the most repeated gene families of the T. cruzi CL Brener genome with 565 gene copies and 136 pseudogenes. Additionally, members of the family are among the largest genes of the parasite [5]. Despite having been discovered more than 30 years ago [6], the role of DGF-1 remains a mystery.
2. The Discovery of the DGF-1 Family
Wincker et al. (1990) [6] discovered the DGF-1 family while working on the genome of the Didelphis marsupialis T. cruzi strain Dm28, reporting 220 members ranging in length from 10 to 12 Kbp and representing about 1% of the parasite’s genome. In Northern blot experiments with pulse-field resolved chromosomes, it was found that the DGF-1 genes were spread throughout the genome and did not have internally repeated sequences. Interestingly, these authors postulated that, given the dispersion of the family and the large gene size, they could participate in inter- and intrachromosomal recombination events, generating chromosomal size polymorphisms. In a second work [8], the same authors registered abundant DGF-1 transcripts in the replicative epimastigote forms of the parasite and sequenced a 10 Kbp gene that was dubbed DGF-1.1. Based on the presence of cysteine rich motifs spread along the gene, they hinted at a potential role of this protein as a receptor, and the presence of two tripeptides RGD suggested potential interactions with the host cell. In DNA hybridization experiments with other trypanosomatids such as Phytomonas sp., Leptomonas samueli, Blastocrithidia sp., Crithidia fasciculata, and Trypanosoma rangeli, these authors reported that the family was exclusive of T. cruzi strains. However, DGF-1 genes were later found in whole genome sequencing of T. rangeli strains Sc-58, Coachi, and M80 [9,10] and more recently in the African stercoraria trypanosomes T. theileri [11] and T. grayi [12].
3. General Molecular Characteristics of DGF-1 Proteins
After the description of the first DGF-1 gene (DGF-1.1) [6], other members of the family were characterized [13,14,15], whose most relevant features are: the presence of eight to nine transmembrane hydrophobic helices at their C-end that might serve as anchors to membranes six epidermal growth factor-1-like (EGF-1), and one EGF-2-like signature regularly spaced approximately 400 aa apart; and lectin binding motifs. There are also integrin-like sequences suggesting a cell surface location [13,14,15] and a possible role either interacting with other cells or as signal transduction receptors. These integrin-like motifs are closer to the primitive integrins (protointegrins) of Saccharomyces cerevisiae, Entamoeba histolytica, Candida albicans, and Dyctiostelium, having in common the RGD tripeptide mentioned above [6,16]. Approximately half of the family members have a canonical signal peptide, and all members lack a GPI anchor site [13]. A topology model of DGF-1.2 inserted in the cell membrane summarizing some features based on sequence homology searches, to which researchers added new information derived from recent advances in structural biology [17,18], is presented in Figure 1. The new structural and biological information revealed that the extracellular part of the protein has a sequence and structural identity with interactive domains such as pectin lyases, phage tail spikes, or receptors associated with virulence factors. The general topology also resembles those of well-studied receptors, with an external cytoplasmic interactive region and an internal cytoplasmic region which may be a signaling transmitting region [19,20].
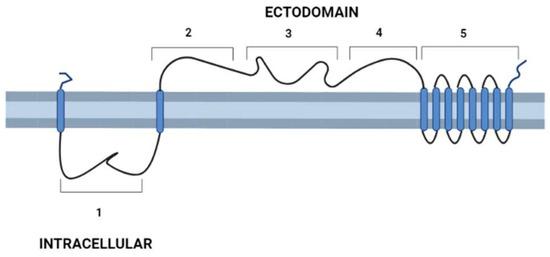
Figure 1. Schematic representation of T. cruzi’s DGF-1.2 protein in the cell membrane. Cell membrane cytoplasmic-extracellular topology predicted by UNIPRO. Numbers mark domains predicted by different structure-prediction software. 1. β-sheet structure (aa 60–140) identity to Human Galectin 7 2–3 (SWISSPRO); pectin-lyase superfamily IPRO11050 (aa 882–987) (InterPro); Regions 2, 3, and 4 with several structural predictions based on sequence identity, structure similarity, and amino acid folding: Complement component C9 receptor (aa 1189–1227) (aa 1584–1622) (SWISS-PRO); Notch ligand (Panther prediction) (aa 1192–1621); pectine lyase-fold (aa 1670–1898) (Alphafold-InterPro); arabinofuranosyltransferase aftd2 from mycobacteria (aa1993–2170) structure prediction confidence 96% (Phyre-2); 4. Threonine-rich region (aa 2800–2927) PROSITE, and structure of tailspike protein gp49 from pseudomonas phage2 lka1 88% confidence (Phyre-2), epitopes from reference [21] are located in this region; 5. Transmembrane helices region DGF1_14, DGF_1.5 (InterPro). Blue cylinders: Transmembrane domains. Graph made with BioRender.
4. DGF-1 Genealogy
In their exhaustive study of the DGF-1 family in the CL Brener strain, Kawashita et al. (2009) [13], based on homology studies and the distribution of putative functional domains, divided the family into two main groups that included most gene members (66 and 51, respectively). The phylogenetic analysis also suggested that the family might have expanded through gene duplication events, but given the sequence similarity among members, these events were relatively recent or subjected to homogenization events. Also, the presence of parallel edges in the two main groups indicates that the family has experienced reticulate events such as recombination or gene conversion [13]. Two members of the family were clearly outgroups, namely XP 807429 (non-Esmeraldo-like haplotype type I group) and XP 816326 (Esmeraldo-like haplotype type II group), both lacking the pectin-lyase motif but having an immunoglobulin-like fold and a cysteine-proteinase inhibitor domain not present in the rest of the family. As mentioned before, members of the DGF-1 family have been found in T. rangeli, T. theliri, and T. gayi, and using genomic data, they were clustered in a distinct phylogenetic group [11].
Jackson et al. (2016) [22] proposed that DGF-1 genes appear to be innovations from more ancestral genes present in free-living organisms included in the genus Bodo, generically designated as Bodonin genes. Bodonin is defined as a multicopy gene family of transmembrane glycoproteins. The typical Bodonin gene has seven transmembrane anchors at the C-terminal, preceded by an extracellular domain, which in turn is preceded by an intracellular domain. The overall organization of the Bonodin and DGF-1.2 genes is shown in Figure 2.
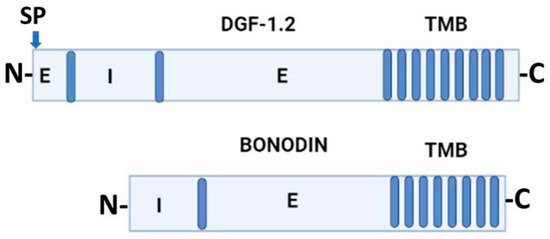
Figure 2. Schematic representation of T. cruzi DGF-1.2 protein and Bodo saltans Bonodin gene [17]. Barrels represent transmembrane regions; SP, signal peptide; N and C protein terminals; I, intracellular regions; E, extracellular regions. Graph made with BioRender.
5. Chromosomal Distribution and DGF-1 Copy Numbers
DGF-1 gene organization was also revealed by Olivares et al. (2000) [23], who, while analyzing sequences of large T. cruzi BAC recombinants, reported DGF-1 members intermingling with the L1Tc retrotransposon. Later, Kim et al. (2005) [14] reported DGF-1 copies in the subtelomeric regions of BAC-telomere recombinants of the CLBrener strain. Interestingly, as in the case of Olivares et al. (2000) [23], the DGF-1 copies were surrounded by genes and pseudogenes of the trans-sialidases, retrotransposon hotspot sequence (RHS) families, and retrotransposon elements. Despite this seemingly unstable environment, the DGF-1 copies were uninterrupted ORFs.
The subtelomeric location of DGF-1 has been confirmed in different T. cruzi strains [24,25,26], but there is a considerable variation in copy numbers among strains [24]. Berna et al. (2018) [26] found that DGF-1 genes were often clustered with other repeated families in what they called “disrupted chromosomal regions”. Following this idea, then, subtelomeres are part of these regions and are likely to be involved in T. cruzi chromosomal gene variation dynamics [14,25,27]. A very interesting observation made by de Bezerra de Araujo et al. [28] is the presence of chromosomal replication origins in subtelomeric DGF-1 members; this location could produce frontal collisions with the transcriptional machinery running in the direction of the telomere and cause chromosomal instability [29], chromosomal breaks, and recombination events leading to genetic variability.
This entry is adapted from the peer-reviewed paper 10.3390/pathogens12020292
This entry is offline, you can click here to edit this entry!
