Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Immunology
The camelid-derived single chain antibody (sdAb), also termed VHH or nanobody, is a unique, functional heavy (H)-chain antibody (HCAb). In contrast to conventional antibodies, sdAb is a unique antibody fragment consisting of a heavy-chain variable domain. It lacks light chains and a first constant domain (CH1). With a small molecular weight of only 12~15 kDa, sdAb has a similar antigen-binding affinity to conventional Abs but a higher solubility, which exerts unique advantages for the recognition and binding of functional, versatile, target-specific antigen fragments.
- sdAb
- HCAb
- nanobody
- VHH
1. Introduction
Antibodies, also called immunoglobulins (Ig), are at the core of an adaptive immune system. They have the ability to recognize and eliminate specific foreign substances from the body. Due to a natural ability to bind to target antigens, antibodies demonstrate great value in biomedicine and drug development [1,2]. To improve the clinical therapeutic efficacy, increase antigen recognition specificity, eliminate the immunogenicity and improve the affinity and stability of antibodies, monoclonal antibodies (mAb) were designed and produced. Since 1975, a significant amount of effort has been invested in monoclonal antibodies. Many monoclonal-antibody-targeted drugs were generated and used in biomedical research, disease treatment and clinical diagnosis [3]. As common biological macromolecules, monoclonal antibodies have had a particularly important pharmaceutical, commercial value over the past several decades [4], such as acting as an antibody–drug conjugate in the combination of CD147 monoclonal antibodies (CD147 mAb) and camptothecin polyphosphoester nanoparticles to effectively and precisely target hepatocellular carcinoma cells, improving the clinical, therapeutic effect on tumors [5]. However, at present, it is also very mature to obtain monoclonal antibodies for clinical disease treatment through phage library construction technology. Monoclonal antibodies are still macromolecules and have a complex internal structure. Multiple factors, such as a susceptibility to variable region folding, a low macromolecular permeability in tissues, and a high host immunogenicity, greatly hinder the development and application of antibody medicines [4].
A natural, small, functional antibody known as a heavy-chain antibody (HCAb) was first reported in camelid serum in 1993 [6]. Unlike conventional antibodies with a heterotetrametric structure, camelid-derived HCAb is devoid of light-chain polypeptides and lacks the first constant domain (CH1) in heavy-chains. Remarkably, the antigen-binding fragment in HCAbs contains only one single-variable domain. This domain is termed VHH and is also known as a single-domain antibody (sdAb) or nanobody (Nb) [7]. Uniquely, the strict, monomeric state of these nanobodies provides the ability to recognize and bind antigens independently after cloning and expression in vitro [8], such as the simplified, monomeric VHH-Fc antibodies [9,10] and small-scale, secretory VHH expression in Saccharomyces cerevisiae [11]. With a smaller biomolecule in the size range of 12~15 kDa and a higher affinity and stability, nanobodies, as promising agents in antibody engineering, have successfully attracted the attention of many biologists. For instance, humanized VHHs [12,13,14], the recombinant variable domains of heavy-chain-only antibodies that are made using a recombinant gene technique [15], have been used in cancer-targeting therapy. Obviously, the appearance of nano-antibody targeting drugs can greatly promote the development of antibody drugs in the field of tumor therapy when compared with traditional mAbs. CAR-T therapy brought a breakthrough in immuno-cancer treatment. Excitingly, research has demonstrated that VHH-based CAR-Ts disrupt the major limitations posed by the single-chain fragment variable (scFv) of an mAb; these limitations cover anti-idiotypic responses and pre-matured scFv aggregation, resulting in antigen-independent CAR-T exhaustion [16]. To summarize, nanobodies form the basis of a wide variety of applications, such as biological research, biomaterial development, medical diagnosis, immunotherapy and clinical trials.
2. Existing Forms of HCAb in Camelid Serum
Conventional IgGs are tetramers consisting of two heavy-chains and two light chains (Figure 1A). The heavy-chain of an IgG includes three constant domains: CH1, CH2 and CH3, and a variable domain, the VH region. The light chain of an IgG contains a variable domain, VL, and a constant domain, CL (Figure 1A). Heavy-chains and light chains are linked by disulfide bonds between the VH and CH1. The variable regions, VH and VL, each contain three hypervariable, complementarity-determining regions (CDRs) that assemble to help form the correct antigen-binding site. Scientists are dedicated to manufacturing small, recombinant antibodies such as the single-chain fragment variable (scFv), which was generated by the concatenation of VH and VL via a linker oligopeptide using genetic engineering in vitro (Figure 1A) [17,18,19], and the fragment of antigen binding (Fab), which contains a light chain and the first two domains (VH and CH1 domain) of a heavy-chain, linked by hinges or disulfide bonds (Figure 1A) [20]. While the small, recombinant antibody has great potential for use in many fields, its development is limited by the insufficient affinity of VH and VL for antigens.
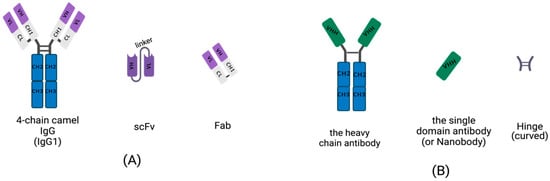
Figure 1. Schematic diagram for the primary structure of a conventional antibody, heavy-chain antibody (HCAb), nanobody, and small, recombinant antibody (This figure was created with BioRender.com, accessed on 27 March 2022). (A) A 4-chain camel IgG (IgG1), a class-conventional antibody in mammal serum, which consists of two light chains and two heavy-chains. The single light chain contains a variable region, VL, and a constant region, CL. The single heavy-chain contains a variable region, CL, and three constant regions, CH1, CH2 and CH3. The scFv is composed of a VH and VL pair-linked by an oligopeptide. A Fab fragment is formed by the first two domains of heavy-chain and light chain. (B) The single chain of HCAb contains two constant regions, CH2 and CH3 and a variable region, VHH, linked by curved hinge. VHH domain owns the smallest intact antigen-binding fragments, also known as single-domain antibodies (sdAbs) or nanobodies.
Homodimeric antibodies are different from the conventional, heterotetrametric antibodies that naturally exist in the serum of camelids, including camels, alpacas and vicugnas. The homodimeric antibody has only two heavy-chains and is devoid of light chains; it is named HCAb (Figure 1B) [21]. Both conventional, 4-chain IgGs and HCAbs are present in camelid serum and possess the abilities of antigen-specific recognition and binding. Camelid HCAb is charactered by the absence of light chains and the lack of the CH1 in the heavy-chain. Therefore, the single-variable domains easily form a propensity for concave surfaces as active sites or receptor-binding pockets. HCAbs contain only two constant regions, CH2 and CH3, and a variable region, VHH, in each chain (Figure 1B). The terminal domain of a VHH is to engage with an antigen. This is the smallest functional, antigen-binding fragment in an HCAb, also known as an sdAb or a nanobody (Figure 1B).
Biochemical Features of the sdAb
4.1. Stability
Chemical inductions, such as a high concentration of guanidine hydrochloride (GuHcl) and urea, can lead to the reversible unfolding of antigens specifically bound to the CDR region and labile, refolded protein molecules. These can be achieved using far ultraviolet CD spectroscopy and surface plasmon resonance (SPR) for monitoring. In addition, the synergistic unfolding of the nanobody fragments was also observed by pressure induction. The above studies illustrate that the VHH fragment of HCAbs in camelid serum has a remarkably high conformational stability [35,36].
SdAbs can perform functions under high temperature exposure due to their ability to refold following heat denaturation. The VHH fragment of the HCAb in camel serum has a distinct thermo-reversible stability profile [37,38]. Most of the denatured VHH-R2 fragment population can be refolded at an extremely high temperature. These VHH-R2 molecules, induced by antigen–antibody production, are in an unfolded conformation before binding, indicating that the CDR region of the antigen recognition site in VHH has a non-canonical conformation, making the HCAb fragments fully thermally reversible [39,40]. The VHH fragments selected from the phage display library showed reversible thermal overlap with less aggregation via transient thermal denaturation [41]. Additionally, some efforts can further enhance the thermal stability of sdAbs by raising their melting temperature. Two common methods are: CDR grafting onto known, stable frameworks, and making point mutations to residues that have been identified to enhance stability from highly stable exemplars [42,43]. Notably, the insertion of a non-canonical disulfide bond between framework regions often yields an increase of 10–20 °C in melting temperature without considering the influence on affinity [41]. Evidence showed that the atypical Cys pairs generated in the VHH structure of the HCAbs via the replacement of Cys residues by Leu45 [41] in CDR-FRs produced a non-canonical disulfide linkage between CDR1-CDR3, endowing the VHH fragment with selective stabilization [44]. Additionally, increases in the net negative charge of proteins by chemical modification can increase the resistance of proteins to irreversible inactivation upon exposure to denaturing conditions and decrease the rate of protein aggregation, which is widely used in enzyme improvement [45]. It is also a potential method for improving the stability of a nanobody.
Recently, molecular dynamics (MD) simulations were established to predict mutations affecting the thermal stabilization of sdAbs [46]. This approach estimated the relative stability for a set of sdAbs for which both crystal structures and the melting transition temperature (Tm) were available. Correlations between the fraction of native atomic contacts (denoted by Q) and the experimental Tm values were generated based on computing the Q for the sdAbs at three different temperatures [46]. This approach provided valuable guidance for identifying potential mutation sites that are unlikely to be easily ascertained by any other means, which is useful for modeling to direct t experiments [41].
4.2. Affinity (Solubility)
The hydrophilicity of an antibody’s light-chain surface [47], which are adept at absorbing the hydrophobic surfaces of nonionic surfactants [48], is very important for the development of antibody drugs. The variable heavy-chain domain of human antibodies arises from recombination, and the dispersed proteins re-aggregate under physical conditions such as heat and acid. The characteristics of protein aggregation were studied by analyzing the CDR sequence of the phage display, which is convenient for the research, development, and application of VHs libraries [49]. Furthermore, researchers analyzed the CDR-region amino acid residues through the phage display of a human VH library with a constrained CDR3 loop. This yielded non-aggregating VHs, and the non-aggregative VHs almost existed in the non-canonical CDR1-CDR3 loop at the acid isoelectric point (PI). The PI distribution analysis of the human VH sequence and the Camelidae rearranged VHH sequence revealed that the VHH–acid ratio was higher. Therefore, the design of VHs libraries with non-canonical and non-aggregation intra-CDR disulfide bonds can greatly improve the solubility of the VH domain [50]. In addition, as it is known that proteins behave with minimal solubility at the isoelectric point, increasing the negative charge of the nanobodies in CDR sequences via mutation or modification can also prevent protein aggregation upon denaturation by, for instance, heat or acid [51].
In the VHH of an HCAb, the CDR-H3 region has a larger frequency variation, ranging from 10 to 24 amino acids [52], while the CDR3 fragment of the human VH domain has only 11.6 amino acids [53]. Therefore, compared to conventional Ig antibodies, the variable domain structure of the camelid sdAb has relatively longer CDR3 fragments. As can be seen in the 2.5A resolution representation of the complex crystal structure of the CDR3 region, CDR3s more readily provide antigen-binding sites on their surface grooves, and other parts of CDR3s can also penetrate deeply into the active site of a lysozyme complex. These unique characteristics of the camel VHH fragment determine its higher affinity, or solubility, and have certain anti-aggregation properties [54,55,56].
4.3. Epitope Recognition of Multivalent Bispecific Antibody of Nbs-Based Minimal Size
In recent decades, bispecific antibodies (BsAbs), as intermediate-sized biomolecules, have attracted extensive attention as therapeutic tools in the field of medicine [57,58]. Although BsAbs have excellent pharmacokinetics, they demonstrate extremely low penetration in renal tissue [59]. It is necessary to develop molecular binding mechanisms for soluble, double and trivalent conjugates to improve the retargeting function of effector T cells to tumors [60,61].
The VH and VL domains of the conventional antibody IgG were covered via two independently functioning VHH domains of the HCAb, assembled into bispecific VHH-IgG and combined with a heavy-chain heterodimerization technology to produce monovalent, bispecific, IgG-like antibodies [62]. Pekar’s team used the strand exchange engineering domain (SEED) technology to optimize the heterodimer of the VHH, which was inserted into the Fc backbone to generate trivalent, specific, IgG-like compounds. The generation of multivalent compounds can use ten amino acid linkers to connect the binding sites so that a VHH is engrafted onto a constant region, CH1, while the other VHH-based paratope is replaced on the CH1 region of the light chain, making full use of the biochemical properties of nanobodies without reducing the nanobodies’ affinity to construct a tetravalent, bispecific, IgG-like molecule [63].
Antigen-specific nanobodies are obtained by animal immunization and yeast antibody coupling agent display technology. The exogenous VHH is non-immunogenic [64], and humanized nanobodies are developed by altering the differentially marked amino acids of the VHH fragment [65].
5. Generation of VHH Libraries
The VHH repertoire of the dromedary HCAb is a biological protein molecule, produced through the stimulation of the B cells of immune animals with an antigen. The high-affinity VHH library generated via specifically combining two cognate antigens is also called the immune library. A unique advantage of maturated, in vivo immune libraries is the relatively high quality of VHH sequences to avoid the immunogenicity of antibodies [66,67]. In addition, there are two other types of VHH libraries (or nanobody libraries), i.e., naïve libraries [68] and synthetic libraries [69].
Some researchers immunized camelids with an anti-DNA mouse mAb, which can cause an anti-idiotypic response and activate the humoral immune system, generating VHHs by constructing phage libraries [70]. However, the molecular mimicry approach makes it difficult to obtain syngeneic anti-Ids using anti-idiotypics. Some other studies prepared variable domain fragments of useful, single-domain recombinant antibodies by immunizing llama libraries as an alternative to conventional, monoclonal antibody libraries [71]. Using the immunized alpaca B lymphocytes for VHH libraries, the construction has the unique properties of high titers of intact, single-domain antigen-cognate binders and an ease of affinity. These immunoselected, recombinant nanobody libraries are well expressed. Antigen-binding nanobody (chromobody) technology for the identification and tracking of antigens in living cells is a new and mature tool for the selection and identification of VHH libraries [72,73].
Here, the critical steps in the preparation of an immunized VHH library are descriped. As is shown in Figure 2, camels are immunized to obtain a B lymphocyte blood sample in vivo, and the library is then cloned for display on bacteriophage particles in E. coli [74]. A single-domain VHH library with a minimal, fully engineered, antigen-binding fragment from a camelid and strict-monomer state, heavy-chain-only immunoglobulins with high-quality VHH sequences are responsive to specific antigen binding agents and small library sizes (106–108). Furthermore, affinity-matured, immune nanobody repertoires from these high-standard, immunized, in vivo procedures can better reach pre-specified epitopes and adapt to the immunogenicity of humanized antibodies for the preparation of bioengineered sdAbs [66,75].
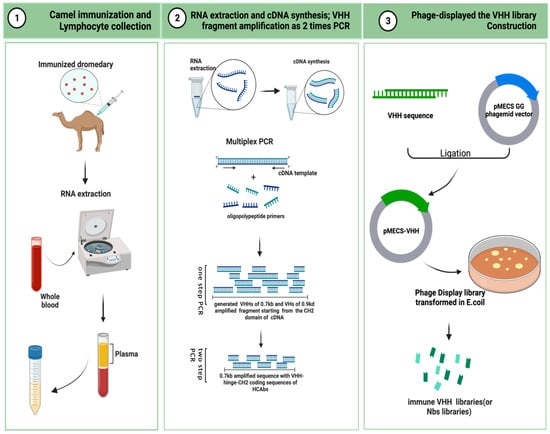
Figure 2. Schematic representation to generate immune VHH libraries (This figure was created with BioRender.com, accessed on 8 August 2022).
5.1. Camel Immunization and Lymphocyte Collection
A healthy adult camel was injected with the antigenic protein of a mixed-compound adjuvant once every two weeks, four to eight times in two months. Camels can use less than approximately 200 mg of mixed adjuvant per injection. The anticoagulated blood of 50 mL from an immunized animal were collected after eight injections. B-lymphocytes were isolated from the immunized camel serum for total RNA extraction, and stored at −80 °C for later use. For long-term storage, the samples were placed in liquid nitrogen [76,77].
5.2. RNA Extraction and cDNA Synthesis: VHH Fragment Amplification from Two-Step PCR [78]
The total RNA was extracted from approximately 107 B-lymphocyte apheresis samples for cDNA synthesis. Oligomeric polypeptide primers were designed based on the single-chain variable domain sequences of the Ig antibodies, and PCR amplification was performed to obtain the VHH gene fragments.
The two-step PCR amplification began from the CH2 domain of an IgG cDNA template. It yielded two distinct PCR states of approximately 0.7 kb and 0.9 kd. The 0.7 kb amplification product was derived from the mRNA coding sequence of the variable domain of the homodimer antibody single chain, which required further agarose gel purification. The purified product was used as a template for PCR amplification again, and only the VHH sequence of the HCAb was obtained.
5.3. Phage Display of the VHH Library Construction [76]
The pMECS GG (a pUC-derived bacteriophage with an F1 responsive source and an IPTG-inducible Plac promoter expression) or another suitable phage display vector were used to ligate the gene fragments of the VHHs to the two-step, amplified PCR products to obtain affinity-matured, robust, recombinant VHH repertoires. The VHH gene sequence ligated to the recombinant vector was again purified and then transformed in E. coli TG1 to generate a small library (107 to 108) of high titer of target-specific binders:s the VHH library.
This entry is adapted from the peer-reviewed paper 10.3390/ijms24044176
This entry is offline, you can click here to edit this entry!
