Metabolomics is the latest trend in the “-omics” sciences, of which technologies are widely used today in all life sciences. Metabolomics gave impetus to the description of biochemical processes that occur in many organisms, search for new biomarkers of disease, and laid the foundation for new clinical laboratory diagnostics. The purpose of this entry is to show how metabolomics is represented in the science, what main research areas were chosen, and to demonstrate the successes and main achievements of scientists in this field.
- metabolomics
- mass spectrometry
- blood
- diagnostics
- aging
- diseases
1. Introduction
Metabolomics is known as a science in which technology platforms are aimed at identifying and quantifying low molecular weight compounds (metabolites). Metabolites are substrates and products of almost all biochemical reactions taking place in the body, play a key role in energy generation, signal transmission in the cell, and carry information about the physiological state of a living organism and ongoing pathological processes [1][2][3]. Metabolomics is a relatively new science and the youngest of the triad of basic “-omics”, including genomics and proteomics, which systematically describe biological objects (Figure 1).
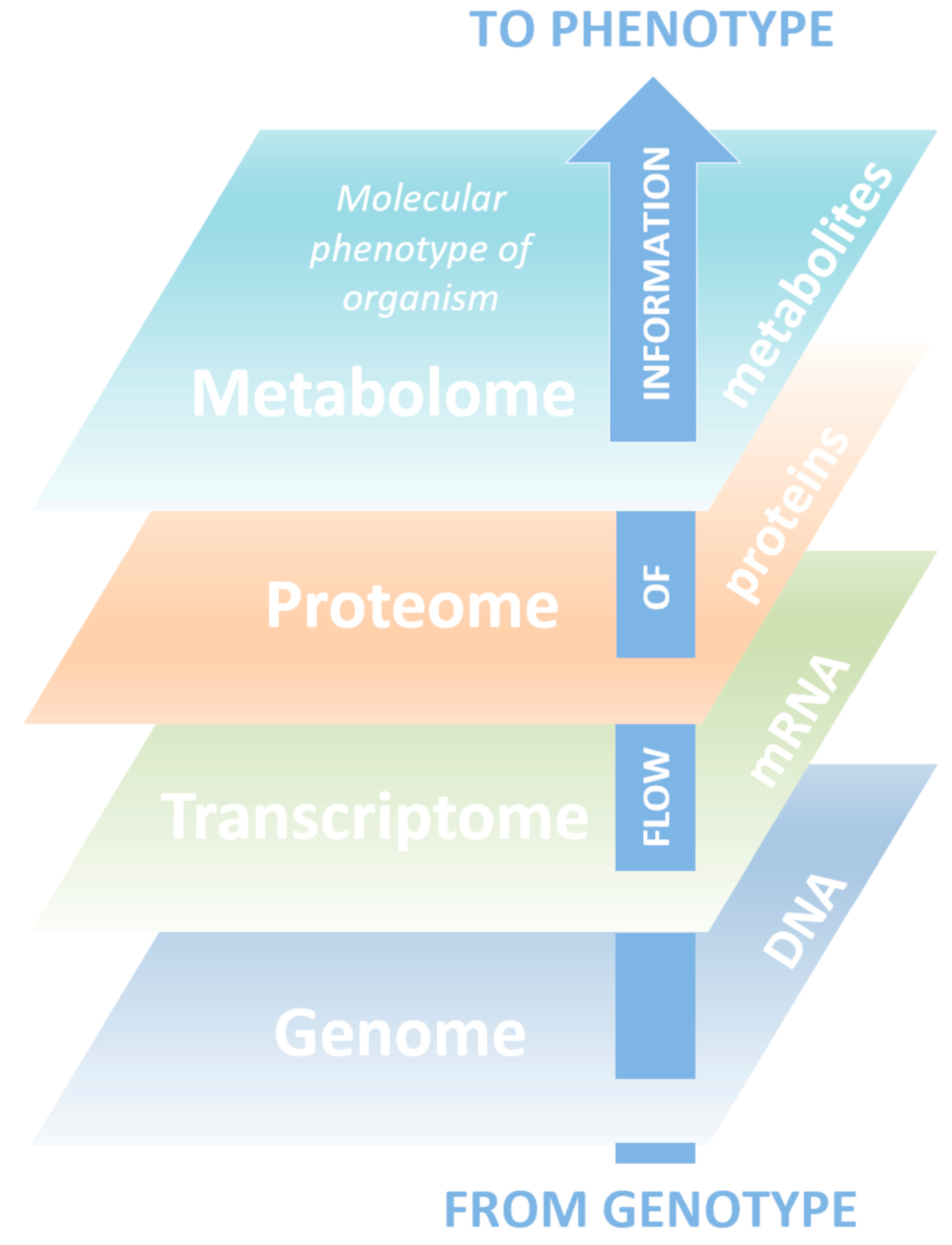
The term “metabolome” was first introduced in 1998, but the main development of metabolomics was observed after 2010 [4], largely due to the use of new, constantly improving high-performance analytical methods and comprehensive bioinformatics data processing [5][6]. Along with other “-omics”, metabolomics research focuses on directions where achievements can bring significant benefits to humanity. Generally, metabolomics studies are able to give a representation of the biochemical processes that underlie the body’s reaction to internal and external stimuli.
2. Current Trends in Metabolomics
Over the past decade, metabolomics has closed an important gap in the post-genomic research [4][7]. Determining the flow of information from the genome to the transcriptome, proteome and, finally, the metabolome allows, for the first time, to comprehensively describe living systems. However, to create models and datasets for accurate modeling, metabolic pathway reconstructions and highly efficient empirical metabolomic research technologies are essential.
A large number of scientists around the world have focused on metabolomics and the research methods and application of metabolomics have been optimized [8]. The Metabolomics Society was created in 2004 as a result of the growing interest in metabolomics. It was established to bring together, under one roof, scientists from the study areas representing metabolite target analysis, metabolic profiling, including metabolic fingerprinting or footprinting, metabolic flux analysis, biochemical modeling, and related bioinformatics fields. In 2007, the Metabolomics Standards Initiative (MSI) formed a general consensus on minimum reporting standards for metabolomics experiments [8]. Until now, all over the world these standards are used to monitor and develop protocols for standardization of the metabolic studies workflow and general analysis approaches [9][10][11][12].
2.1. Diseases Diagnostics
With continuous optimization and improvement of high-tech research methods, metabolomics has become a wide area for the use of “-omics” technologies for medical purposes, the results of which are promising for future implementation [13][14][15]. With the support of the National Cancer Institute (NCI), Food and Drug Administration (FDA), and others, the Institute of Medicine (IOM) has been called upon to define the criteria and procedures for assessing the validity of tests created for the clinic. The IOM appointed the Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials composed of the experts with wide range of knowledge and experience, including experts in “-omics” technologies. The committee presented the results of its activities in the book “Evolution of Translational Omics: Lessons Learned and the Path Forward”, published in 2012 [16]. As a result of the committee’s work, the criteria for preparing of “-omics” tests for clinical trials in humans have been created. The creation of “-omics” tests in accordance with these criteria, in particular, the writing of new protocols for them, reflects the modern understanding of the way to implement these tests into clinical practice. The studies of scientists on the development of laboratory tests for Parkinson’s disease, impaired glucose tolerance, lung cancer and prostate cancer, obesity made a great contribution to this direction [17][18][19][20][21][22][23][24][25][26].
It is worth noting another one initiative recently formed by The Metabolomics Society due to the fact that the study of metabolism at the global or “-omics” level is a rapidly growing field that has the potential to have a profound impact upon medical practice [27]. However, today clinicians utilize only a very small part of the information contained in the metabolome, as they routinely measure only a narrow set of blood chemistry analytes to assess health and disease states [27]. It is expected that “the narrow range of chemical analyses in current use by the medical community today will be replaced in the future by analyses that reveal a far more comprehensive metabolic signature. This signature is expected to describe global biochemical aberrations that reflect patterns of variance in states of wellness, more accurately describe specific diseases and their progression, and greatly aid in differential diagnosis” [27]. The work of scientists dedicated to metabolomic blood analysis, human digital imaging, and label-free data standartization for clinical metabolomics is aimed at supporting this initiative [28][29][30][31][32].
One of the potential uses of metabolomics is the identification of metabolic profiles in body fluids that can predict the effectiveness and toxicity of drugs, the so-called pharmacometabonomics focused on the personalization of drug therapy [33]. The metabolic profile reflects the metabolism of xenobiotics in the body. That is, the metabolic profile contains all information necessary to calculate the effective dose of a drug for a particular patient, taking into account not only the characteristics of the individual pharmacokinetics of the drug, but also the body’s response to it [34][35][36][37].
2.2. Metabolomics of Aging
Today, no one doubts that metabolomics is a promising tool for studying aging, since it is a powerful tool for cataloging changes in the body that occur over time at the molecular level. Aging is a determined process in living organisms, which is characterized by a gradual decrease in physiological activity and reproductive ability, as well as an increase in the frequency of mortality over time. Nevertheless, aging remains one of the most mysterious and not yet fully studied biological phenomena. By measuring numerous small molecules that represent the entire spectrum of metabolic pathways metabolomics can potentially help identify processes that are associated with aging or even lead to it [38][39][40][41][42].
2.3. Diseases Pathogenesis
Metabolomics studies the biochemical and photochemical processes that occur in living organisms and are responsible for the development of diseases. One of the areas of laboratory research is the study of the metabolomic composition of human and laboratory animals (rat, rabbit, calf, fish) tissues, since the development of pathological processes leads to significant changes in the metabolomic composition of the tissue: a decrease or increase in the concentration of various metabolites. Changes in the metabolomic content of tissue are studied by chromatography, mass spectrometry, and nuclear magnetic resonance (NMR) spectroscopy. The main attention is focused on tissues and biological fluids. Using metabolomic information, one can understand the mechanisms of the formation and development of undesirable processes and evaluate the effectiveness of drugs for the prevention and treatment of diseases [43][44][45][46][47].
2.4. Plant Metabolomics
The metabolic processes occurring in plants are widely studied. According to the FAO (Food and Agricultural Organization of the United Nation), potatoes, rice, wheat, corn represent the most important food crop in terms of production. The importance of these plants is difficult to overestimate; it is a valuable source of carbohydrates, antioxidants, and vitamins. In recent years, a large number of studies have focused on the study of the plant metabolome for deciphering the mechanisms responsible for the productivity and accumulation of compounds that determine the taste and nutritional qualities, while maintaining the quality of tubers, plant resistance, etc. [48][49][50]. Complex studies of metabolic diversity using modern chromatographic approaches and high-precision NMR spectroscopy and mass spectrometric detection of individual compounds have revealed the specificity of metabolomic spectra both at the cellular and organism levels under the influence of both internal and external stimuli. Metabolomic approaches are used for phenotyping available lines and varieties of plants, assessing the resistance of plants to environmental factors, and detecting changes in tubers during long-term storage. Metabolomic profiling is widely used to study the differences between genetically modified forms and non-transformed parent plants. In the future, studies of plant metabolome will be able to complement traditional and molecular genetic approaches to breeding to create new lines and varieties that carry valuable features [48][49][50].
This entry is adapted from the peer-reviewed paper 10.3390/biotech9040020
References
- Christian Gieger; Ludwig Geistlinger; Elisabeth Altmaier; Martin Hrabé De Angelis; Florian Kronenberg; Thomas Meitinger; Hans-Werner Mewes; Heinz-Erich Wichmann; Klaus M. Weinberger; Jerzy Adamski; et al. Genetics Meets Metabolomics: A Genome-Wide Association Study of Metabolite Profiles in Human Serum. PLoS Genetics 2008, 4, e1000282, 10.1371/journal.pgen.1000282.
- Caroline H. Johnson; Julijana Ivanisevic; Gary Siuzdak; Metabolomics: beyond biomarkers and towards mechanisms. Nature Reviews Molecular Cell Biology 2016, 17, 451-459, 10.1038/nrm.2016.25.
- Gary J. Patti; Oscar Yanes; Gary Siuzdak; Metabolomics: the apogee of the omics trilogy. Nature Reviews Molecular Cell Biology 2012, 13, 263-269, 10.1038/nrm3314.
- Farhana R. Pinu; Seyed Ali Goldansaz; Jacob Jaine; Translational Metabolomics: Current Challenges and Future Opportunities. Metabolites 2019, 9, 108, 10.3390/metabo9060108.
- Douglas B. Kell; Stephen G. Oliver; The metabolome 18 years on: a concept comes of age. Metabolomics 2016, 12, 1-8, 10.1007/s11306-016-1108-4.
- Richard D. Beger; Warwick B. Dunn; Abbas Bandukwala; Bianca Bethan; David Broadhurst; Clary B. Clish; Surendra Dasari; Leslie Derr; Annie Evans; Steve Fischer; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 1-5, 10.1007/s11306-018-1460-7.
- Qiang Yang; Ai-Hua Zhang; Jian-Hua Miao; Hui Sun; Ying Han; Guang-Li Yan; Fang-Fang Wu; Xi-Jun Wang; Metabolomics biotechnology, applications, and future trends: a systematic review. RSC Advances 2019, 9, 37245-37257, 10.1039/c9ra06697g.
- Lloyd W Sumner; Alexander Amberg; David A. Barrett; Michael H. Beale; Richard D. Beger; Clare A. Daykin; Teresa W.-M. Fan; Oliver Fiehn; Royston Goodacre; Julian Griffin; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211-221, 10.1007/s11306-007-0082-2.
- Petr G. Lokhov; A. I. Archakov; Mass spectrometry methods in metabolomics. Biochemistry (Moscow), Supplement Series B: Biomedical Chemistry 2009, 3, 1-9, 10.1134/s1990750809010016.
- Petr G. Lokhov; Anna A. Voskresenskaya; Oxana P Trifonova; Dmitry L. Maslov; Ekaterina A. Shestakova; Elena E. Balashova; Andrey V. Lisitsa; Prediction of classical clinical chemistry parameters using a direct infusion mass spectrometry. International Journal of Mass Spectrometry 2015, 388, 53-58, 10.1016/j.ijms.2015.08.006.
- Dmitry L. Maslov; Oxana P Trifonova; Elena E. Balashova; Petr G. Lokhov; n-Butylamine for Improving the Efficiency of Untargeted Mass Spectrometry Analysis of Plasma Metabolite Composition. International Journal of Molecular Sciences 2019, 20, 5957, 10.3390/ijms20235957.
- Oxana P Trifonova; Dmitry L. Maslov; Elena E. Balashova; Petr G. Lokhov; Evaluation of Dried Blood Spot Sampling for Clinical Metabolomics: Effects of Different Papers and Sample Storage Stability. Metabolites 2019, 9, 277, 10.3390/metabo9110277.
- Sergei Moshkovskii; Mikhail Pyatnitsky; Petr G. Lokhov; Ancha Baranova; OMICS for Tumor Biomarker Research. Biomarkers in Bone Disease 2015, 1, 3-30, 10.1007/978-94-007-7681-4_14.
- Lisa M McShane; Margaret M. Cavenagh; Tracy G. Lively; David A. Eberhard; William L. Bigbee; P. Mickey Williams; Jill P. Mesirov; Mei-Yin C. Polley; Kelly Y. Kim; James V. Tricoli; et al. Criteria for the use of omics-based predictors in clinical trials. Nature 2013, 502, 317-320, 10.1038/nature12564.
- Patrick M. Bossuyt; Where Are All the New Omics-Based Tests?. Clinical Chemistry 2014, 60, 1256-1257, 10.1373/clinchem.2014.223339.
- Christine M. Micheel, Sharly J. Nass, and Gilbert S. Omenn. Evolution of Translational Omics: Lessons Learned and the Path Forward; Christine M. Micheel, Sharly J. Nass, and Gilbert S. Omenn, Eds.; National Academies Press: Washington, DC, USA, 2012; pp. all.
- Petr G. Lokhov; Maxim I. Dashtiev; Sergey A. Moshkovskii; Alexander I. Archakov; Metabolite profiling of blood plasma of patients with prostate cancer. Metabolomics 2009, 6, 156-163, 10.1007/s11306-009-0187-x.
- Petr G. Lokhov; M. I. Dashtiev; L. V. Bondartsov; A. V. Lisitsa; S. A. Moshkovskii; A. I. Archakov; Metabolic fingerprinting of blood plasma from patients with prostate cancer. Biochemistry (Moscow), Supplement Series B: Biomedical Chemistry 2010, 4, 37-41, 10.1134/s1990750810010051.
- Petr G. Lokhov; Oxana P Trifonova; Dmitry L. Maslov; Elena E. Balashova; Alexander I. Archakov; Ekaterina A. Shestakova; Marina V. Shestakova; Ivan I. Dedov; Diagnosing Impaired Glucose Tolerance Using Direct Infusion Mass Spectrometry of Blood Plasma. PLOS ONE 2014, 9, e105343, 10.1371/journal.pone.0105343.
- Petr G. Lokhov; Oleg N. Kharybin; Alexander I. Archakov; Diagnosis of lung cancer based on direct-infusion electrospray mass spectrometry of blood plasma metabolites. International Journal of Mass Spectrometry 2012, 309, 200-205, 10.1016/j.ijms.2011.10.002.
- Petr G. Lokhov; Oxana P. Trifonova; Dmitry L. Maslov; Alexander I. Archakov; Blood plasma metabolites and the risk of developing lung cancer in Russia. European Journal of Cancer Prevention 2013, 22, 335-341, 10.1097/cej.0b013e32835b3898.
- Elena E. Balashova; Petr G. Lokhov; Dmitry L. Maslov; Oxana P. Trifonova; Diana M. Khasanova; Zuleykha A. Zalyalova; Razina R. Nigmatullina; Alexander I. Archakov; Michael V. Ugrumov; Plasma Metabolome Signature in Patients with Early-stage Parkinson Disease. Current Metabolomics 2017, 6, 75-82, 10.2174/2213235x05666170221161735.
- Petr G. Lokhov; Elena E. Balashova; Oxana P Trifonova; Dmitry L. Maslov; Elena A. Ponomarenko; Alexander Archakov; Mass Spectrometry-Based Metabolomics Analysis of Obese Patients’ Blood Plasma. International Journal of Molecular Sciences 2020, 21, 568, 10.3390/ijms21020568.
- Petr G. Lokhov; Elena E. Balashova; Anna A. Voskresenskaya; Oxana P. Trifonova; Dmitry L. Maslov; Alexander I. Archakov; Mass spectrometric signatures of the blood plasma metabolome for disease diagnostics. Biomedical Reports 2015, 4, 122-126, 10.3892/br.2015.548.
- Petr G. Lokhov; Oxana P. Trifonova; Dmitry L. Maslov; Steven Lichtenberg; Elena E. Balashova; Diagnosis of Parkinson’s Disease by A Metabolomics-Based Laboratory-Developed Test (LDT). Diagnostics 2020, 10, 332, 10.3390/diagnostics10050332.
- Oxana P. Trifonova; Dmitry L. Maslov; Elena E. Balashova; Guzel R. Urazgildeeva; Denis A. Abaimov; Ekaterina Yu. Fedotova; Vsevolod V. Poleschuk; Sergey N. Illarioshkin; Petr G. Lokhov; Parkinson’s Disease: Available Clinical and Promising Omics Tests for Diagnostics, Disease Risk Assessment, and Pharmacotherapy Personalization. Diagnostics 2020, 10, 339, 10.3390/diagnostics10050339.
- Richard D. Beger; for “Precision Medicine and Pharmacometabolomics Task Group”-Metabolomics Society Initiative; Warwick Dunn; Michael A. Schmidt; Steven S. Gross; Jennifer A. Kirwan; Marta Cascante; Lorraine Brennan; David S. Wishart; Matej Oresic; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 1-15, 10.1007/s11306-016-1094-6.
- P.G. Lokhov; A.V. Lisitsa; A.I. Archakov; Metabolomic blood test: purpose, implementation and interpretation of data. Biomeditsinskaya Khimiya 2017, 63, 232-240, 10.18097/pbmc20176303232.
- Petr G. Lokhov; Dmitri L. Maslov; Oleg N. Kharibin; Elena E. Balashova; Evgeny N. Nikolaev; Label-free data standardization for clinical metabolomics. BioData Mining 2017, 10, 1-12, 10.1186/s13040-017-0132-x.
- Elena E Balashova; Petr G Lokhov; Elena A Ponomarenko; Sergey S Markin; Andrey V Lisitsa; Alexander I Archakov; Metabolomic diagnostics and human digital image. Personalized Medicine 2019, 16, 133-144, 10.2217/pme-2018-0066.
- Oxana P Trifonova; Petr G. Lokhov; A. I. Archakov; Metabolic profiling of human blood. Biochemistry (Moscow), Supplement Series B: Biomedical Chemistry 2013, 7, 179-186, 10.1134/s1990750813030128.
- Andrey V. Lisitsa; Elena A. Ponomarenko; P. G. Lokhov; A. I. Archakov; Postgenomic Medicine: Alternative to Biomarkers. Annals of the Russian academy of medical sciences 2016, 71, 255-260, 10.15690/vramn647.
- D.L. Maslov; E.E. Balashova; P.G. Lokhov; A.I. Archakov; Pharmacometabonomics – the novel way to personalized drug therapy. Biomeditsinskaya Khimiya 2017, 63, 115-123, 10.18097/pbmc20176302115.
- Dmitry L. Maslov; Elena E. Balashova; Oxana P Trifonova; Petr G. Lokhov; Metabolomics-based Approach to Pharmacotherapy Personalization: Advantages and Limitations. Current Pharmacogenomics and Personalized Medicine 2019, 16, 192-198, 10.2174/1875692116666181008144905.
- Elena E. Balashova; Dmitry L. Maslov; Petr G. Lokhov; A Metabolomics Approach to Pharmacotherapy Personalization. Journal of Personalized Medicine 2018, 8, 28, 10.3390/jpm8030028.
- P.G. Lokhov; D.L. Maslov; O.P. Trifonova; E.E. Balashova; A.I. Archakov; Mass spectrometry of blood low-molecular fraction as a method for unification of therapeutic drug monitoring. Biomeditsinskaya Khimiya 2014, 60, 201-216, 10.18097/pbmc20146002201.
- P.G. Lokhov; D.L. Maslov; E.E. Balashova; O.P. Trifonova; N.V. Medvedeva; T.I. Torkhovskaya; O.M. Ipatova; A.I. Archakov; P.P. Malyshev; V.V. Kukharchuk; et al. Mass spectrometry analysis of blood plasma lipidome as method of disease diagnostics, evuation of effectiveness and optimization of drug therapy. Biomeditsinskaya Khimiya 2015, 61, 7-18, 10.18097/pbmc20156101007.
- Oxana P Trifonova; Dmitry L. Maslov; Anton N. Mikhailov; Konstantin V Zolotarev; Kirill V. Nakhod; Valeriya I. Nakhod; Nataliya F. Belyaeva; Marina V. Mikhailova; Petr G. Lokhov; Alexander Archakov; et al. Comparative Analysis of the Blood Plasma Metabolome of Negligible, Gradual and Rapidly Ageing Fishes. Fishes 2018, 3, 46, 10.3390/fishes3040046.
- Dmitry L. Maslov; Oxana P Trifonova; Anton N. Mikhailov; Konstantin V Zolotarev; Kirill V. Nakhod; Valeriya I. Nakhod; Nataliya F. Belyaeva; Marina V. Mikhailova; Petr G. Lokhov; Alexander Archakov; et al. Comparative Analysis of Skeletal Muscle Metabolites of Fish with Various Rates of Aging. Fishes 2019, 4, 25, 10.3390/fishes4020025.
- Nikolaos Psychogios; David D. Hau; Jun Peng; An Chi Guo; Rupasri Mandal; Souhaila Bouatra; Igor Sinelnikov; Ramanarayan Krishnamurthy; Roman Eisner; Bijaya Gautam; et al. The Human Serum Metabolome. PLOS ONE 2011, 6, e16957, 10.1371/journal.pone.0016957.
- Robert J. Mishur; Shane L. Rea; Applications of mass spectrometry to metabolomics and metabonomics: Detection of biomarkers of aging and of age-related diseases. Mass Spectrometry Reviews 2012, 31, 70-95, 10.1002/mas.20338.
- Yuqing He; Zheng Yu; Ina Giegling; Lu Xie; Annette M Hartmann; Cornelia Prehn; Jerzy Adamski; Rene S Kahn; Yun Li; Thomas Illig; et al. Schizophrenia shows a unique metabolomics signature in plasma. Translational Psychiatry 2012, 2, e149-e149, 10.1038/tp.2012.76.
- Anthony Au; Metabolomics and Lipidomics of Ischemic Stroke. Advances in Applied Microbiology 2018, 85, 31-69, 10.1016/bs.acc.2018.02.002.
- Ashish Kumar; Biswapriya Biswavas Misra; Challenges and Opportunities in Cancer Metabolomics. PROTEOMICS 2019, 19, e1900042, 10.1002/pmic.201900042.
- Olga A. Snytnikova; Lyudmila V. Yanshole; Igor A. Iskakov; Vadim V. Yanshole; Valery V. Chernykh; Denis A. Stepakov; Vladimir P. Novoselov; Yuri P. Tsentalovich; Quantitative metabolomic analysis of the human cornea and aqueous humor. Metabolomics 2017, 13, 152, 10.1007/s11306-017-1281-0.
- Olga A. Snytnikova; Anastasiya A. Khlichkina; Lyudmila V. Yanshole; Vadim V. Yanshole; Igor A. Iskakov; Elena V. Egorova; Denis A. Stepakov; Vladimir P. Novoselov; Yuri P. Tsentalovich; Metabolomics of the human aqueous humor. Metabolomics 2016, 13, 5, 10.1007/s11306-016-1144-0.
- Vadim V. Yanshole; Lyudmila V. Yanshole; Olga A. Snytnikova; Yuri P. Tsentalovich; Quantitative metabolomic analysis of changes in the lens and aqueous humor under development of age-related nuclear cataract. Metabolomics 2019, 15, 29, 10.1007/s11306-019-1495-4.
- Jun Hong; Litao Yang; Dabing Zhang; Jianxin Shi; Plant Metabolomics: An Indispensable System Biology Tool for Plant Science. International Journal of Molecular Sciences 2016, 17, 767, 10.3390/ijms17060767.
- Leonardo Perez De Souza; Saleh Alseekh; Thomas Naake; Alisdair R. Fernie; Mass Spectrometry‐Based Untargeted Plant Metabolomics. Current Protocols in Plant Biology 2019, 4, e20100, 10.1002/cppb.20100.
- R. K. Puzanskiy; V. V. Yemelyanov; T. A. Gavrilenko; M. F. Shishova; The perspectives of metabolomic studies of potato plants. Russian Journal of Genetics: Applied Research 2016, 7, 744-756, 10.1134/s207905971707005x.
