Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
DNA cytosine methylation is a principal epigenetic mechanism underlying transcription during development and aging. Growing evidence suggests that DNA methylation plays a critical role in brain function, including neurogenesis, neuronal differentiation, synaptogenesis, learning, and memory.
- DNA methylation
- transcriptional regulation
- neurogenesis
1. Introduction
The brain is the most important organ that serves as the center of the nervous system in mammals. The function of the brain is to control and coordinate a wide variety of actions and reactions, especially thought, memory, and emotion in primates. The development and aging of the brain are complex processes that depend on multiple layers of precise regulation. During aging, the brain shrinks and changes structurally and is predisposed to neurodegenerative diseases. Alzheimer’s disease (AD), Parkinson’s disease (PD), and Huntington’s disease (HD) are common neurodegenerative diseases that cause progressive loss of neuronal function and lead to cognitive impairment. Epigenetic mechanisms play essential roles in brain function by regulating gene expression [1][2][3]. As a critical epigenetic modification, DNA methylation affects transcriptional activity by recruiting or inhibiting the binding of transcription factors to DNA. Many brain disorders display altered gene expression, and accumulating evidence implicates dynamic DNA methylation in altered gene expression and pathological processes in the brain [4][5]. Dysregulation of the epigenome has been reported in neurodevelopmental and neurodegenerative diseases [6].
DNA methylation is a heritable epigenetic mechanism linked to gene expression and the regulation of biological processes [7]. In mammals, 5-methylcytosine (5mC) is formed by DNA methyltransferases (DNMTs) transferring a methyl group from S-adenosylmethionine (SAM) to cytosine at 5-position (Figure 1A). 5mC occurs mainly in the CpG dinucleotides of the mammalian genome. A total of 70–80% of CpGs are modified by 5mC in the human genome [8]. During development and cellular differentiation, the initial establishment of 5mC patterns, termed de novo methylation, relies predominantly on the activity of both DNMT3a and DNMT3b [9], whereas DNMT1 performs a maintenance function over the course of DNA replication to ensure the propagation of 5mC patterns [10].
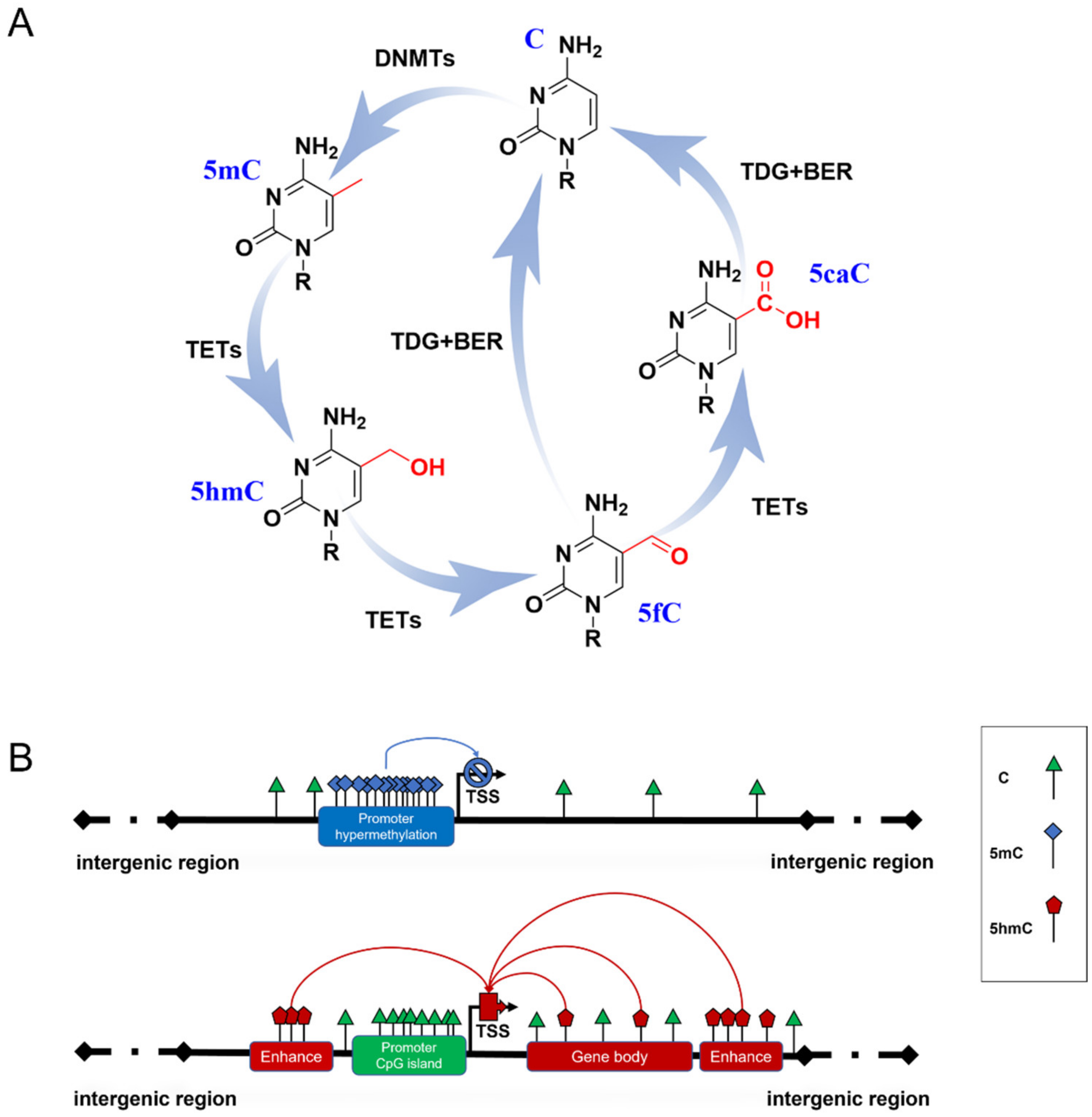
Figure 1. DNA methylation formation and regulation in gene expression. (A) The cycle of DNA methylation. 5mC is formed from C by the catalyzation of DNMTs; then, it can be hydroxylated to 5hmC and further oxidized to 5fC and 5CaC by TET enzymes. 5fC and 5caC are recognized by thymine DNA glycosylase (TDG), which triggers the base excision repair (BER) process, leading to the conversion to unmethylated cytosine. (B) Possible role of 5mC and 5hmC in gene expression. Hypermethylated 5mC at CpG islands of gene promoters generally inhibits gene transcription (blue line). 5hmC of gene enhancers and gene bodies up-regulates gene expression (red line).
5mC has been termed the fifth base of the human genome because it plays a key role in inhibiting transcriptional activities (Figure 1B) [11]. A typical situation is the X chromosome inactivation in placental mammals, where 5mC shuts down the transcription of most genes on one of the female’s X chromosomes to compensate for the different dosages of the X chromosome in males and females [12]. However, some studies have reported that several methylation sites are related to gene transcriptional activation [13][14], which may be due to the dynamic binding of transcription factors to certain methylated DNA. 5mC patterns are subject to orderly changes during mammalian development and aging. Neurogenesis proceeds from embryonic stages to the adult brain [15][16]. Dynamic DNA methylation indeed contributes to embryonic and adult neurogenesis, including stem cell maintenance and proliferation, neuronal differentiation and maturation, fate interpretation, and synaptogenesis. Gain or loss of 5mC impacts neuronal development and brain function [17][18]. Growing evidence supports Horvath’s epigenetic clock theory, which is based on 5mC patterns of specific sites across multiple tissues and provides an estimator for predicting chronological age [19]. During aging, an overall genome-wide decreased 5mC (global hypomethylation) has been reported, and regional hypermethylation within the CpG islands of specific gene promoters has been observed [20][21]. Abnormal patterns of 5mC commonly disrupt transcriptional regulation and lead to various diseases, such as cancer and neurodegenerative diseases.
Another family, named Ten-eleven translocation (TET) dioxygenases, is responsible for catalyzing the iterative oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) on DNA. The TET family in mammals contains Tet1, Tet2, and Tet3, which share a homology catalytic domain at their C terminal. 5fC and 5caC are then specifically recognized by thymine DNA glycosylase to trigger the base excision repair (BER) process, leading to their final conversion to unmethylated cytosine (Figure 1A) [22][23]. Originally, this process was thought to be just an active DNA demethylation with 5hmC, 5fC, and 5caC as transient intermediates. Proteome-wide profiling showed that these modifications may recruit different transcriptional regulators (as readers) to regulate gene expression (Figure 1B) [24][25]. This suggests that novel forms of DNA methylation provide potential regulatory mechanisms for genome functions. Due to the limitations of detection methods for 5fC and 5caC, most studies have focused on the biological functions of 5hmC.
Large differences in 5hmC levels in different tissues have been reported, ranging from 0.03% to 0.69% [26]. 5hmC is typically enriched in active genes, promoters, transcription factor-binding sites, and brain-specific enhancers but is absent in intergenic regions [27][28]. At present, a great number of studies have confirmed that 5hmC generally promotes gene expression. Therefore, 5hmC is considered to be a stable epigenetic marker and is thus called the sixth base of DNA [29][30]. In mammals, 5hmC is the most abundant in the central nervous system (CNS), where mature neurons appear to be the major contributors to 5hmC. 5hmC levels in CNS are approximately 10-fold higher than in embryonic stem cells (ESCs) and 3-fold higher than in peripheral tissues, especially those enriched in Purkinje neurons [26][27][31]. This suggests that high levels of 5hmC are essential for proper neurodevelopment and contribute to the neuropathology of neurodegenerative diseases [5][32].
In the pathogenesis of various neurodegenerative diseases, the silencing or activation of multiple genes has been characterized by hypomethylation or hypermethylation and abnormal regulation of enzymes related to DNA methylation formation or removal. As epigenetic modifications are invertible processes, this provides insights into therapeutic intervention by correcting the aberrant epigenetic status to achieve epigenetic balance. DNA methylation emerges as a promising target for the selection of therapeutic approaches. To date, DNMT inhibitors (DNMTi) are widely used in preclinical and clinical research, among which 5-Azacytidine (5-Aza-CR) and 5-aza-2′-deoxycitidine (5-Aza-CdR) have been approved by the Food and Drug Administration (FDA) in the United States [33][34]. Depending on the interaction of epigenetic modifications, the new therapy has also been tried in clinical applications by combining DNMTi and HDACi (histone deacetylases inhibitors) [35][36][37]. Further understanding of the fundamental mechanisms of epigenetic regulation in neuronal development and diseases could promote therapeutic applications.
2. Dynamic Changes in DNA Methylation in Brain Development
5mC plays a prominent role in the normal development of organisms. Abnormal changes in 5mC patterns may lead to developmental disorders, such as intellectual disability, global developmental delays, movement disorders, and growth abnormalities [38]. At early developmental stages, neural stem cells generate all cortical neurons. 5mC alterations at the promoters of genes, such as Slit1, Bdnf, Wnt3, Esrrb, and Tcl1, that are involved in brain development and neural differentiation may result in transcriptional changes in those genes and, therefore, influence neural differentiation [39]. Cytosine methylation occurs primarily at CpG dinucleotides but is also found at non-CpGs sites. There is a significant association between gene silencing and non-CpG methylation [40]. Notably, non-CpG methylation is a conserved DNA modification in the human neuronal genome that is implicated in the regulation of genes related to neuronal differentiation, synaptogenesis, and function [38]. Unlike CpG-methylation, non-CpG methylation is established after the maturation of neurons in human and mouse brains. In humans, 5mC levels at certain loci that are responsible for the growth and development of CNS increase significantly after birth and participate in neuronal differentiation—a dramatic change that can also persist throughout the whole life [41][42]. Interestingly, mature neurons never undergo mitosis, which means that although 5mC cannot be passively eliminated during cell division, it can still be actively abolished by TETs [43]. Therefore, 5mC is dynamic and plays a role in neurogenesis. Similarly, DNA methylation is involved in the regulation of astrocyte differentiation and maturation. A potential impact on sexual differentiation in the developing mammalian brain has been implicated, in which 5mC is required for the masculinization or feminization of the brain [44][45].
The dynamics of 5mC depend on a series of enzymes, including DNMT1 for methylation maintenance and DNMT3a/b for de novo methylation. DNMT1 is a highly conserved DNA methyltransferase in mice and humans that maintains the DNA methylation pattern faithfully during cell division [46]. In mice, DNMT3a is highly expressed in neural stem and progenitor cells in the developing cerebral cortex and neurons and oligodendrocytes (but with very low expression in astrocytes) [47][48]. DNMT3b is expressed only in the ventricular zone during the early stages of embryonic development [17]. Mice deleted of Dnmt3a (Dnmt3a-/-) can survive for four weeks, but Dnmt1 or Dnmt3b homozygous deletion (Dnmt1-/-, Dnmt3b-/-) resulted in embryonic or postnatal lethality [9][49]. Conditional mutant mice that lack Dnmt1 or Dnmt3a in forebrain excitatory neurons exhibited learning and memory impairment and deregulated gene expression in neurons. These findings revealed the roles of Dnmt1 and Dnmt3a in maintaining DNA methylation and modulating neuronal gene expression in adult neurons [50]. Furthermore, Dnmt3b is essential for de novo methylation during development, and a lack of Dnmt3b led to abnormal development of the rostral neural tube [9].
5hmC mappings of mice and humans showed that genome-wide 5hmC levels in brain tissues are significantly higher than in other tissues, reaching more than 10-fold [51][52][53]. 5hmC levels are increased markedly from the early postnatal period until adulthood, suggesting a strong correlation between elevated 5hmC and expression levels of genes important for normal neural development and activity [28][53]. Tet1 and Tet2 are highly expressed in embryonic stem cells ESCs [51]. However, Tet3 expression levels were barely detectable in ESCs but rapidly increased during neuronal differentiation [54]. Several studies have shown that the regulation of the expression of 5hmC and TET family enzymes is important for neurogenesis and neurodevelopment. Tet1/2/3 triple knockout in ESCs compromised neural differentiation and manifested as developmental defects during gastrulation in early mouse embryos [55][56]. The loss of Tet2 contributed to enhancer hypomethylation by oxidizing 5mC and delayed the timing of transcriptome reprogramming, which influenced the differentiation of ESCs to neural progenitor cells (NPCs) [57]. In Tet3-knockdown of NPCs, a dramatic genome-wide loss of 5hmC has been reported, which led to a de-repressing of pluripotency-associated genes such as Oct4 and Nanog. These findings indicated that Tet3 plays a critical role in NPC specification, maintenance, and terminal differentiation [39][54][58].
During neurodevelopmental stages, Tet1 deficiency resulted in stage-dependent defects during the transition from NPCs to oligodendrocyte progenitor cells (OPCs), which, in turn, affected the formation of mature myelinating oligodendrocytes (OLs) and remyelination in the mouse brain [52][59]. The inhibition of Tet1/Tet3 impaired the branching of GCs dendrites and gene expression related to axon guidance, dendrite outgrowth, and ion channel functions [26][60]. Tet3 was required for regulating excitatory and inhibitory synaptic transmission [61]. In clinical observations, human TET3 deficiency led to the common phenotypic features of intellectual disability and/or overall developmental delays [62]. 5hmC was preferentially enriched in genes related to synapse-associated functions in human and mouse brains [63]. Genome-wide 5hmC profiling data revealed a unique pattern in the cerebellum during brain development in rhesus monkeys [64]. It has been demonstrated that 5hmC levels increase sharply during cerebellar granule cell (GC) development, which is consistent with the accumulation of 5hmC in the cerebellum [60]. Taken together, all the above-mentioned studies suggest that 5hmC is involved in the epigenetic regulation of normal physiological function and neuronal circuits in the cerebellum.
In conclusion, it is clear that the establishment of and dynamic changes in 5mC/5hmC are involved in neuronal development and differentiation in a cell- and region-specific manner, evidenced by the inactivation of methylation-associated enzymes that can result in developmental anomalies and lethality.
This entry is adapted from the peer-reviewed paper 10.3390/biology12020152
References
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232.
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21.
- Robertson, K.; Wolffe, A.P. DNA methylation in health and disease. Nat. Rev. Genet. 2000, 1, 11–19.
- Singleton, A.; Hardy, J.; Armstrong, M.J.; Jin, Y.; Allen, E.G.; Jin, P. Progress in the genetic analysis of Parkinson’s disease. Hum. Mol. Genet. 2019, 28, R241–R253.
- MacArthur, I.C.; Dawlaty, M.M. TET Enzymes and 5-Hydroxymethylcytosine in Neural Progenitor Cell Biology and Neurodevelopment. Front. Cell Dev. Biol. 2021, 9, 645335.
- Younesian, S.; Yousefi, A.-M.; Momeny, M.; Ghaffari, S.H.; Bashash, D. The DNA Methylation in Neurological Diseases. Cells 2022, 11, 3439.
- Santos, K.; Mazzola, T.; Carvalho, H. The prima donna of epigenetics: The regulation of gene expression by DNA methylation. Braz. J. Med. Biol. Res. 2005, 38, 1531–1541.
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476.
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257.
- Vertino, P.M.; Sekowski, J.A.; Coll, J.M.; Applegreen, N.; Han, S.; Hickey, R.J.; Malkas, L.H. DNMT1 is a Component of a Multiprotein DNA Replication Complex. Cell Cycle 2002, 1, 416–423.
- Wu, C.-T.; Morris, J.R. Genes, Genetics, and Epigenetics: A Correspondence. Science 2001, 293, 1103–1105.
- Wutz, A. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011, 12, 542–553.
- Halpern, K.B.; Vana, T.; Walker, M.D. Paradoxical Role of DNA Methylation in Activation of FoxA2 Gene Expression during Endoderm Development. J. Biol. Chem. 2014, 289, 23882–23892.
- Niesen, M.I.; Osborne, A.R.; Yang, H.; Rastogi, S.; Chellappan, S.; Cheng, J.Q.; Boss, J.M.; Blanck, G. Activation of a Methylated Promoter Mediated by a Sequence-specific DNA-binding Protein, RFX. J. Biol. Chem. 2005, 280, 38914–38922.
- Ming, G.-L.; Song, H. Adult Neurogenesis in the Mammalian Brain: Significant Answers and Significant Questions. Neuron 2011, 70, 687–702.
- Götz, M.; Huttner, W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 777–788.
- Wang, Z.; Tang, B.; He, Y.; Jin, P. DNA methylation dynamics in neurogenesis. Epigenomics 2016, 8, 401–414.
- Zocher, S.; Overall, R.W.; Berdugo-Vega, G.; Rund, N.; Karasinsky, A.; Adusumilli, V.S.; Steinhauer, C.; Scheibenstock, S.; Händler, K.; Schultze, J.L.; et al. De novo DNA methylation controls neuronal maturation during adult hippocampal neurogenesis. EMBO J. 2021, 40, e107100.
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115, Erratum in Genome Biol. 2015, 16, 96.
- Vlaming, H.; van Leeuwen, F. Crosstalk between aging and the epigenome. Epigenomics 2012, 4, 5–7.
- Fuke, C.; Shimabukuro, M.; Petronis, A.; Sugimoto, J.; Oda, T.; Miura, K.; Miyazaki, T.; Ogura, C.; Okazaki, Y.; Jinno, Y. Age Related Changes in 5-methylcytosine Content in Human Peripheral Leukocytes and Placentas: An HPLC-based Study. Ann. Hum. Genet. 2004, 68, 196–204.
- Wu, H.; Zhang, Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011, 25, 2436–2452.
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133.
- Spruijt, C.G.; Gnerlich, F.; Smits, A.H.; Pfaffeneder, T.; Jansen, P.W.T.C.; Bauer, C.; Munzel, M.; Wagner, M.; Muller, M.; Khan, F.; et al. Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell 2013, 152, 1146–1159.
- Bai, L.; Yang, G.; Qin, Z.; Lyu, J.; Wang, Y.; Feng, J.; Liu, M.; Gong, T.; Li, X.; Li, Z.; et al. Proteome-Wide Profiling of Readers for DNA Modification. Adv. Sci. 2021, 8, 2101426.
- Globisch, D.; Münzel, M.; Müller, M.; Michalakis, S.; Wagner, M.; Koch, S.; Brückl, T.; Biel, M.; Carell, T. Tissue Distribution of 5-Hydroxymethylcytosine and Search for Active Demethylation Intermediates. PLoS ONE 2010, 5, e15367.
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.-X.; Wu, H.; Dai, Q.; Irier, H.; Upadhyay, A.K.; Gearing, M.; Levey, A.I.; et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011, 14, 1607–1616.
- Hahn, M.A.; Qiu, R.; Wu, X.; Li, A.X.; Zhang, H.; Wang, J.; Jui, J.; Jin, S.-G.; Jiang, Y.; Pfeifer, G.P.; et al. Dynamics of 5-Hydroxymethylcytosine and Chromatin Marks in Mammalian Neurogenesis. Cell Rep. 2013, 3, 291–300.
- Muenzel, M.; Globisch, D.; Brückl, T.; Wagner, M.; Welzmiller, V.; Michalakis, S.; Müller, M.; Biel, M.; Carell, T. Quantification of the Sixth DNA Base Hydroxymethylcytosine in the Brain. Angew. Chem. Int. Ed. 2010, 49, 5375–5377.
- Bachman, M.; Uribe-Lewis, S.; Yang, X.; Williams, M.; Murrell, A.; Balasubramanian, S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014, 6, 1049–1055.
- Kriaucionis, S.; Heintz, N. The Nuclear DNA Base 5-Hydroxymethylcytosine is Present in Purkinje Neurons and the Brain. Science 2009, 324, 929–930.
- Antunes, C.; Sousa, N.; Pinto, L.; Marques, C.J. TET enzymes in neurophysiology and brain function. Neurosci. Biobehav. Rev. 2019, 102, 337–344.
- Kuo, H.K.; Griffith, J.D.; Kreuzer, K.N. 5-Azacytidine–Induced Methyltransferase-DNA Adducts Block DNA Replication In vivo. Cancer Res. 2007, 67, 8248–8254.
- Stresemann, C.; Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13.
- Soriano, A.O.; Yang, H.; Faderl, S.; Estrov, Z.; Giles, F.; Ravandi, F.; Cortes, J.; Wierda, W.G.; Ouzounian, S.; Quezada, A.; et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood 2007, 110, 2302–2308.
- Voso, M.T.; Santini, V.; Finelli, C.; Musto, P.; Pogliani, E.; Angelucci, E.; Fioritoni, G.; Alimena, G.; Maurillo, L.; Cortelezzi, A.; et al. Valproic Acid at Therapeutic Plasma Levels May Increase 5-Azacytidine Efficacy in Higher Risk Myelodysplastic Syndromes. Clin. Cancer Res. 2009, 15, 5002–5007.
- Bruyer, A.; Maes, K.; Herviou, L.; Kassambara, A.; Seckinger, A.; Cartron, G.; Rème, T.; Robert, N.; Requirand, G.; Boireau, S.; et al. DNMTi/HDACi combined epigenetic targeted treatment induces reprogramming of myeloma cells in the direction of normal plasma cells. Br. J. Cancer 2018, 118, 1062–1073.
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global Epigenomic Reconfiguration During Mammalian Brain Development. Science 2013, 341, 1237905.
- Santiago, M.; Antunes, C.; Guedes, M.; Iacovino, M.; Kyba, M.; Reik, W.; Sousa, N.; Pinto, L.; Branco, M.R.; Marques, C.J. Tet3 regulates cellular identity and DNA methylation in neural progenitor cells. Cell. Mol. Life Sci. 2019, 77, 2871–2883.
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2013, 17, 215–222.
- Siegmund, K.D.; Connor, C.M.; Campan, M.; Long, T.I.; Weisenberger, D.J.; Biniszkiewicz, D.; Jaenisch, R.; Laird, P.W.; Akbarian, S. DNA Methylation in the Human Cerebral Cortex Is Dynamically Regulated throughout the Life Span and Involves Differentiated Neurons. PLoS ONE 2007, 2, e895.
- Fan, G.; Martinowich, K.; Chin, M.H.; He, F.; Fouse, S.D.; Hutnick, L.; Hattori, D.; Ge, W.; Shen, Y.; Wu, H.; et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 2005, 132, 3345–3356.
- He, F.; Wu, H.; Zhou, L.; Lin, Q.; Cheng, Y.; Sun, Y.E. Tet2-mediated epigenetic drive for astrocyte differentiation from embryonic neural stem cells. Cell Death Discov. 2020, 6, 30.
- Nugent, B.M.; Wright, C.L.; Shetty, A.; Hodes, G.; Lenz, K.M.; Mahurkar, A.; Russo, S.; Devine, S.E.; McCarthy, M.M. Brain feminization requires active repression of masculinization via DNA methylation. Nat. Neurosci. 2015, 18, 690–697.
- Cisternas, C.D.; Cortes, L.R.; Bruggeman, E.C.; Yao, B.; Forger, N.G. Developmental changes and sex differences in DNA methylation and demethylation in hypothalamic regions of the mouse brain. Epigenetics 2019, 15, 72–84.
- Ye, F.; Kong, X.; Zhang, H.; Liu, Y.; Shao, Z.; Jin, J.; Cai, Y.; Zhang, R.; Li, L.; Zhang, Y.W.; et al. Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1. ACS Chem. Biol. 2018, 13, 772–781.
- Moyon, S.; Huynh, J.L.; Dutta, D.; Zhang, F.; Ma, D.; Yoo, S.; Lawrence, R.; Wegner, M.; John, G.R.; Emery, B.; et al. Functional Characterization of DNA Methylation in the Oligodendrocyte Lineage. Cell Rep. 2016, 15, 748–760.
- Feng, J.; Chang, H.; Li, E.; Fan, G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005, 79, 734–746.
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992, 69, 915–926.
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.; Fan, G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430.
- Koh, K.P.; Yabuuchi, A.; Rao, S.; Huang, Y.; Cunniff, K.; Nardone, J.; Laiho, A.; Tahiliani, M.; Sommer, C.A.; Mostoslavsky, G.; et al. Tet1 and Tet2 Regulate 5-Hydroxymethylcytosine Production and Cell Lineage Specification in Mouse Embryonic Stem Cells. Cell Stem Cell 2011, 8, 200–213.
- Zhang, M.; Wang, J.; Zhang, K.; Lu, G.; Liu, Y.; Ren, K.; Wang, W.; Xin, D.; Xu, L.; Mao, H.; et al. Ten-eleven translocation 1 mediated-DNA hydroxymethylation is required for myelination and remyelination in the mouse brain. Nat. Commun. 2021, 12, 5091.
- Jin, S.-G.; Wu, X.; Li, A.X.; Pfeifer, G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011, 39, 5015–5024.
- Li, T.; Yang, D.; Li, J.; Tang, Y.; Yang, J.; Le, W. Critical Role of Tet3 in Neural Progenitor Cell Maintenance and Terminal Differentiation. Mol. Neurobiol. 2014, 51, 142–154.
- Dai, H.-Q.; Wang, B.-A.; Yang, L.; Chen, J.-J.; Zhu, G.-C.; Sun, M.-L.; Ge, H.; Wang, R.; Chapman, D.L.; Tang, F.; et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty–Nodal signalling. Nature 2016, 538, 528–532.
- Dawlaty, M.M.; Breiling, A.; Le, T.; Barrasa, M.I.; Raddatz, G.; Gao, Q.; Powell, B.E.; Cheng, A.W.; Faull, K.F.; Lyko, F.; et al. Loss of Tet Enzymes Compromises Proper Differentiation of Embryonic Stem Cells. Dev. Cell 2014, 29, 102–111.
- Hon, G.C.; Song, C.-X.; Du, T.; Jin, F.; Selvaraj, S.; Lee, A.Y.; Yen, C.-A.; Ye, Z.; Mao, S.-Q.; Wang, B.-A.; et al. 5mC Oxidation by Tet2 Modulates Enhancer Activity and Timing of Transcriptome Reprogramming during Differentiation. Mol. Cell 2014, 56, 286–297.
- Morris-Blanco, K.C.; Chokkalla, A.K.; Bertogliat, M.J.; Vemuganti, R. TET3 regulates DNA hydroxymethylation of neuroprotective genes following focal ischemia. J. Cereb. Blood Flow Metab. 2020, 41, 590–603.
- Moyon, S.; Frawley, R.; Marechal, D.; Huang, D.; Marshall-Phelps, K.L.H.; Kegel, L.; Bøstrand, S.M.K.; Sadowski, B.; Jiang, Y.-H.; Lyons, D.A.; et al. TET1-mediated DNA hydroxymethylation regulates adult remyelination in mice. Nat. Commun. 2021, 12, 3359.
- Zhu, X.; Girardo, D.; Govek, E.-E.; John, K.; Mellén, M.; Tamayo, P.; Mesirov, J.P.; Hatten, M.E. Role of Tet1/3 Genes and Chromatin Remodeling Genes in Cerebellar Circuit Formation. Neuron 2015, 89, 100–112.
- Wang, L.; Li, M.-Y.; Qu, C.; Miao, W.-Y.; Yin, Q.; Liao, J.; Cao, H.-T.; Huang, M.; Wang, K.; Zuo, E.; et al. CRISPR-Cas9-mediated genome editing in one blastomere of two-cell embryos reveals a novel Tet3 function in regulating neocortical development. Cell Res. 2017, 27, 815–829.
- Levy, M.A.; Beck, D.B.; Metcalfe, K.; Douzgou, S.; Sithambaram, S.; Cottrell, T.; Ansar, M.; Kerkhof, J.; Mignot, C.; Nougues, M.-C.; et al. Deficiency of TET3 leads to a genome-wide DNA hypermethylation episignature in human whole blood. NPJ Genom. Med. 2021, 6, 92.
- Khare, T.; Pai, S.; Koncevicius, K.; Pal, M.; Kriukiene, E.; Liutkeviciute, Z.; Irimia, M.; Jia, P.; Ptak, C.; Xia, M.; et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 2012, 19, 1037–1043.
- Xu, Y.; Zhong, L.; Wei, H.; Li, Y.; Xie, J.; Xie, L.; Chen, X.; Guo, X.; Yin, P.; Li, S.; et al. Brain Region- and Age-Dependent 5-Hydroxymethylcytosine Activity in the Non-Human Primate. Front. Aging Neurosci. 2022, 14, 934224.
This entry is offline, you can click here to edit this entry!
