Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Polyamines (PAs) are positively charged amines that are present in all organisms. In addition to their functions specific to growth and development, they are involved in responding to various biotic and abiotic stress tolerance functions. The appropriate concentration of PA in the cell is maintained by a delicate balance between the catabolism and anabolism of PAs, which is primarily driven by two enzymes, namely diamine oxidase and polyamine oxidase (PAO).
- polyamine
- abiotic stress
- enzymes
1. Introduction
Polyamines (PAs) are small aliphatic amines with four to ten carbon chain lengths and are ubiquitously present across the kingdoms, from prokaryotic to eukaryotic organisms. Polyamines may exist in multiple forms, such as free-polyamines, covalently conjugated, or non-covalently conjugated (NCC-PAs) forms [1]. The largest pool of PAs is constituted by free polyamines conjugated with phenolic compounds, such as hydroxycinnamic acid, coumaric acid, caffeic acid, or ferulic acid via amide linkage [2][3][4]. In the physiological state, the free PAs exist as fully protonated and positively charged, and hence make a complex with macromolecules, such as nucleic acids, proteins, or lignin via ionic interaction or hydrogen bonding [5]. The most common polyamines in higher plants are putrescine (Put, a di-amine), spermidine (Spd, a tri-amine), and spermine (Spm, a tetra-amine) [3][6][7]. In contrast, in lower plants, like algae and mosses, the unusual PAs, namely norspermidine (NorSpd) and norspermine (NorSpm), constitute the bulk of PAs [8]. Structurally, NorSpd and NorSpm are similar to their more commonly present PA siblings Spd and Spm respectively, except they have one methyl group less in the carbon chain [9][10]. Lately, these unusual polyamines have also been identified in low concentrations in higher plants like Medicago sativa [8][11][12], Arabidopsis thaliana [13], and Zea mays [14]. Another tetra-amine, thermospermine (T-Spm), has been identified both in the lower plant, a diatom (Thalassiosira pseudonana), and in the higher plant A. thaliana [13]. Another secondary diamine similar to Put is cadaverine, which has been reported in Glycine max seedlings [15]. Studies pertaining to PAs such as T-Spm, NorSpd, NorSpm, and cadaverine are still scarce in the literature.
Traditionally, there are three major PAs present in plants: Put, Spm and Spd. However, with more and more data being made available, a fourth PA, namely T-Spm, has also been added to this category. All the PAs have been implicated to have both some common and specific functions. In plants, PAs have been suggested to be involved in a wide range of functions, starting from embryogenesis to flowering and senescence [6][7][16]. Their role in biotic and abiotic stress tolerance has also been documented [3][16][17]. T-Spm, which has been demonstrated to be required for stem elongation in A. thaliana, has also been considered a major PA in higher plants [14][18], while the secondary diamine cadaverine has been reported to be required for root growth in G. max seedlings [15].
PA concentrations in cells fluctuate, and they are governed by a dynamic balance of anabolism and catabolism. Polyamine oxidases (PAOs) play a significant role in PA metabolism and they are therefore of much importance in maintaining the cellular pool of polyamines. PAOs are primarily present in the cytosol and apoplast; however, lately, their peroxisomal localization has also been reported in various plant species. Peroxisomes are an important organelle for abiotic stress responses.
2. Catabolism
The two important enzymes involved in polyamine catabolism are diamine oxidase (DAO) and polyamine oxidase (PAO) [4][19][20]. DAO uses Cu2+ and pyridoxal phosphate as cofactors. It acts upon Put and converts it to 4-aminobutanal with concomitant production of H2O2 and NH3. The 4-aminobutanal is acted upon by the pyrrolinedehydrogenase (PYRR-DH) enzyme and is converted to γ-aminobutyric acid (GABA), followed by conversion to Krebs cycle intermediate, succinate. In comparison to monocots, dicot plants contain higher amounts of DAOs; however, their encoding genes have been cloned from very few plant species [20].
PAO is a flavin adenine dinucleotide (FAD)-dependent enzyme and catalyzes the oxidative deamination of PAs at both the secondary amino groups [19][20]. Unlike DAOs, they have been found to remain present in monocots at high levels [21][22]. PAO enzymes are of two types: terminal catabolism (TC) and back conversion (BC) type. TC type leads to the breakdown of PAs into corresponding aldehydes: 4-aminobutanal and N-(3-aminopropyl)-4-aminobutanal, for Spd and Spm, respectively, along with concomitant production of 1,3-diaminopropane and hydrogen peroxide (H2O2), while the BC-type PAO leads to conversion of tetramine to triamine, and in certain circumstances, of triamine to diamine, leading to an increase in the cellular concentration of PAs [19][23][24][25][26]. The H2O2, which is produced as a byproduct of PA catabolism, has been demonstrated to act as a second messenger in biotic and abiotic stress signal transduction pathways [27][28] (Figure 1). It also affects the closure of stomata mediated by abscisic acid [20][29][30]. It has also been speculated that PAs lead to the accumulation of another second messenger, nitric oxide [31], which has also been deemed necessary for plant growth and development aside from its involvement in biotic and abiotic stress signaling [32].
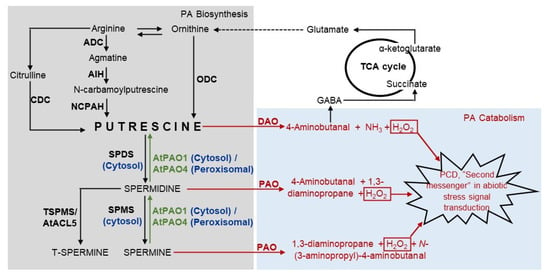
Figure 1. Diagrammatic representation of PA catabolism, anabolism, and their links with the TCA cycle. Grey and blue highlighted boxes represent biosynthesis and catabolism, respectively. Enzymes are represented by bold text. Green texts and arrows represent back conversions. Red texts and arrows represent catabolism. Dotted arrow represents the multi-step conversion. ACL—ACAULIS, ADC—arginine decarboxylase, AIH—agmatine iminohydrolase, CDC—citrulline decarboxylase, DAO—diamine oxidase, GABA—γ-aminobutyric acid, NCPAH—N-carbamoylputrescine amidohydrolase, ODC—ornithine decarboxylase, PAO—polyamine oxidase, PCD—programmed cell death, SPDS—spermidine synthase, SPMS—spermine synthase, TCA—tricarboxylic acid, TSPMS—thermospermine synthase.
3. Biosynthesis
The diamine, Put, is the central compound of PA biosynthesis. In plants, Put is synthesized from two different precursors—ornithine and arginine. Ornithine is converted to Put by the enzyme ornithine decarboxylase (ODC) in a single-step reaction [33][34]. Arginine is converted to Put in a three-step enzymatic reaction, where arginine decarboxylase (ADC) converts arginine to agmatine and carbon dioxide. In the second step, agmatine is converted to N-carbamoylputrescine (NCPA) and ammonia by the enzyme agmatine iminohydrolase (AIH). In the last step the N-carbamolylputrescine amidohydrolase (NCPAH) hydrolyses N-carbamoylputrescine to Put, CO2 and NH3 [34]. This is the primary Put biosynthesis pathway in plants [35][36]. There lies another alternate pathway, where arginine is converted to Put via an intermediate, citrulline, by the enzyme citrulline decarboxylase (CDC) [37][38][39]. The biosynthesis of Put via citrulline is limited in occurrence and has been reported in Sesamum indicum plants only [40]. It has also been observed that the gene ODC has been lost from A. thaliana and many other members of Brassicaceae during the course of evolution [41], suggesting that the ODC-dependent pathway may not be absolutely necessary for normal growth and development [40]. The diamine Put is converted to triamine Spd by the enzyme spermidine synthase, which has been found to be localized in cytosolic fractions [42]. The latter is further converted into tetra-amines Spm and T-Spm by spermine synthase (SPMS) and thermospermine synthase (T-SPMS), respectively, reviewed in [33]. These enzymes catalyze the addition of a fourth amine group. In A. thaliana, ACAULIS5 (ACL5) has been demonstrated to be a thermospermine synthase ortholog, which synthesizes T-Spm from Spd [13][14][43] (Figure 1).
The BC-type PAOs also contribute to the accumulation of the cellular PA pool. The recombinant AtPAO1 has been found to catalyze the back conversion of tetramine Spm and NorSpm to triamine Spd and NorSpd, respectively [44]. In the case of rice, all the three peroxisomal PAOs (OsPAO3, OsPAO4, and OsPAO5) and one cytosolic isoform OsPAO1, carry out PA back conversion from Spm and T-Spm to Spd and Spd to Put [19][45]. The BC-type PAO has also been reported in the lower plant, Selaginella lepidophylla, where it (SelPAO) catalyzes the back conversion of Spm and T-Spm to Spd and NorSpd, respectively. Usually, NorSpd is synthesized from 1,3-diaminopropane (DAP) by the action of aminopropyl transferase (APT). SelPAO synthesis of NorSpd from T-Spm reveals a novel pathway for NorSpd synthesis [8]. From the back-conversion property of the PAO, it may be envisioned that PAO enzymes play a crucial role in maintaining the cellular concentration of polyamines as they are involved both in the catabolism and anabolism of PA, thereby regulating the PAO enzymes, which could be instrumental in the polyamine-dependent stress adaption of plants.
This entry is adapted from the peer-reviewed paper 10.3390/plants12030652
References
- Gholami, M.; Fakhari, A.R.; Ghanati, F. Selective Regulation of Nicotine and Polyamines Biosynthesis in Tobacco Cells by Enantiomers of Ornithine. Chirality 2012, 25, 22–27.
- Martin-Tanguy, J. Conjugated polyamines and reproductive development: Biochemical, molecular and physiological approaches. Physiol. Plant 2010, 100, 675–688.
- Kusano, T.; Berberich, T.; Tateda, C.; Takahashi, Y. Polyamines: Essential factors for growth and survival. Planta 2008, 228, 367–381.
- Bassard, J.-E.; Ullmann, P.; Bernier, F.; Werck-Reichhart, D. Phenolamides: Bridging polyamines to the phenolic metabolism. Phytochemistry 2010, 71, 1808–1824.
- Igarashi, K.; Kashiwagi, K. Modulation of protein synthesis by polyamines. IUBMB Life 2015, 67, 160–169.
- Alcázar, R.; Marco, F.; Cuevas, J.C.; Patron, M.; Ferrando, A.; Carrasco, P.; Tiburcio, A.F.; Altabella, T. Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett. 2006, 28, 1867–1876.
- Groppa, M.D.; Benavides, M.P. Polyamines and abiotic stress: Recent advances. Amino Acids 2007, 34, 35–45.
- Sagor, G.H.M.; Inoue, M.; Kim, D.W.; Kojima, S.; Niitsu, M.; Berberich, T.; Kusano, T. The polyamine oxidase from lycophyte Selaginella lepidophylla (SelPAO5), unlike that of angiosperms, back-converts thermospermine to norspermidine. FEBS Lett. 2015, 589, 3071–3078.
- Fincato, P.; Moschou, P.; Spedaletti, V.; Tavazza, R.; Angelini, R.; Federico, R.; Roubelakis-Angelakis, K.A.; Tavladoraki, P. Functional diversity inside the Arabidopsis polyamine oxidase gene family. J. Exp. Bot. 2010, 62, 1155–1168.
- Michael, A.J. Polyamines in Eukaryotes, Bacteria, and Archaea. J. Biol. Chem. 2016, 291, 14896–14903.
- Liu, T.; Kim, N.W.; Niitsu, M.; Berberich, T.; Kusano, T. POLYAMINE OXIDASE 1 from rice (Oryza sativa) is a functional ortholog of Arabidopsis POLYAMINE OXIDASE 5. Plant Signal. Behav. 2014, 9, e29773.
- Fincato, P.; Moschou, P.; Ahou, A.; Angelini, R.; Roubelakis-Angelakis, K.A.; Federico, R.; Tavladoraki, P. The members of Arabidopsis thaliana PAO gene family exhibit distinct tissue- and organ-specific expression pattern during seedling growth and flower development. Amino Acids 2011, 42, 831–841.
- Knott, J.M.; Romer, P.; Sumper, M. Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 2007, 581, 3081–3086.
- Takano, A.; Kakehi, J.-I.; Takahashi, T. Thermospermine is Not a Minor Polyamine in the Plant Kingdom. Plant Cell Physiol. 2012, 53, 606–616.
- Gamarnik, A.; Frydman, R.B. Cadaverine, an Essential Diamine for the Normal Root Development of Germinating Soybean (Glycine max) Seeds. Plant Physiol. 1991, 97, 778–785.
- Takahashi, Y.; Cong, R.; Sagor, G.H.M.; Niitsu, M.; Berberich, T.; Kusano, T. Characterization of five polyamine oxidase isoforms in Arabidopsis thaliana. Plant Cell Rep. 2010, 29, 955–965.
- Yoda, H.; Hiroi, Y.; Sano, H. Polyamine Oxidase Is One of the Key Elements for Oxidative Burst to Induce Programmed Cell Death in Tobacco Cultured Cells. Plant Physiol. 2006, 142, 193–206.
- Kakehi, J.-I.; Kuwashiro, Y.; Niitsu, M.; Takahashi, T. Thermospermine is Required for Stem Elongation in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1342–1349.
- Ono, Y.; Kim, D.W.; Watanabe, K.; Sasaki, A.; Niitsu, M.; Berberich, T.; Kusano, T.; Takahashi, Y. Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids 2011, 42, 867–876.
- Jaana, V.; Riina, M.; Komlan, A.; Marko, S.; Johanna, K.; Esa, L.; Hely, H.; Outi, S.; Tytti, S. Thermospermine Synthase (ACL5) and Diamine Oxidase (DAO) Expression Is Needed for Zygotic Embryogenesis and Vascular Development in Scots Pine. Front. Plant Sci. 2019, 10, 1600.
- Takahashi, Y.; Ono, K.; Akamine, Y.; Asano, T.; Ezaki, M.; Mouri, I. Highly-expressed polyamine oxidases catalyze polyamine back conversion in Brachypodium distachyon. J. Plant Res. 2017, 131, 341–348.
- Hao, Y.; Huang, B.; Jia, D.; Mann, T.; Jiang, X.; Qiu, Y.; Niitsu, M.; Berberich, T.; Kusano, T.; Liu, T. Identification of seven polyamine oxidase genes in tomato (Solanum lycopersicum L.) and their expression profiles under physiological and various stress conditions. J. Plant Physiol. 2018, 228, 1–11.
- Cervelli, M.; Cona, A.; Angelini, R.; Polticelli, F.; Federico, R.; Mariottini, P. A barley polyamine oxidase isoform with distinct structural features and subcellular localization. JBIC J. Biol. Inorg. Chem. 2001, 268, 3816–3830.
- Cervelli, M.; Di Caro, O.; Di Penta, A.; Angelini, R.; Federico, R.; Vitale, A.; Mariottini, P. A novel C-terminal sequence from barley polyamine oxidase is a vacuolar sorting signal. Plant J. 2004, 40, 410–418.
- Cervelli, M.; Bianchi, M.; Cona, A.; Crosatti, C.; Stanca, M.; Angelini, R.; Federico, R.; Mariottini, P. Barley polyamine oxidase isoforms 1 and 2, a peculiar case of gene duplication. FEBS J. 2006, 273, 3990–4002.
- Liu, T.; Kim, D.W.; Niitsu, M.; Berberich, T.; Kusano, T. Oryza sativa polyamine oxidase 1 back-converts tetraamines, spermine and thermospermine, to spermidine. Plant Cell Rep. 2013, 33, 143–151.
- Freitas, V.S.; Miranda, R.D.S.; Costa, J.H.; De Oliveira, D.F.; Paula, S.D.O.; Miguel, E.D.C.; Freire, R.S.; Prisco, J.T.; Gomes-Filho, E. Ethylene triggers salt tolerance in maize genotypes by modulating polyamine catabolism enzymes associated with H2O2 production. Environ. Exp. Bot. 2018, 145, 75–86.
- Mellidou, I.; Karamanoli, K.; Beris, D.; Haralampidis, K.; Constantinidou, H.-I.A.; Roubelakis-Angelakis, K.A. Underexpression of apoplastic polyamine oxidase improves thermotolerance in Nicotiana tabacum. J. Plant Physiol. 2017, 218, 171–174.
- Ni Tun, N.; Santa-Catarina, C.; Begum, T.; Silveira, V.; Handro, W.; Floh, E.I.S.; Scherer, G.F.E. Polyamines Induce Rapid Biosynthesis of Nitric Oxide (NO) in Arabidopsis thaliana Seedlings. Plant Cell Physiol. 2006, 47, 346–354.
- An, Z.F.; Jing, W.; Liu, Y.L.; Zhang, W.H. Hydrogen peroxide generated by copper amine oxidase is involved in abscisic acid-induced stomatal closure in Vicia faba. J. Exp. Bot. 2008, 59, 815–825.
- Pál, M.; Szalai, G.; Janda, T. Speculation: Polyamines are important in abiotic stress signaling. Plant Sci. 2015, 237, 16–23.
- Agurla, S.; Gayatri, G.; Raghavendra, A.S. Polyamines increase nitric oxide and reactive oxygen species in guard cells of Arabidopsis thaliana during stomatal closure. Protoplasma 2018, 255, 153–162.
- Mattoo, A.K.; Minocha, S.C.; Minocha, R.; Handa, A.K. Polyamines and cellular metabolism in plants: Transgenic approaches reveal different responses to diamine putrescine versus higher polyamines spermidine and spermine. Amino Acids 2009, 38, 405–413.
- Wuddineh, W.; Minocha, R.; Minocha, S.C. Polyamines in the Context of Metabolic Networks. Polyam. Methods Mol. Biol. 2017, 1694, 1–23.
- Docimo, T.; Reichelt, M.; Schneider, B.; Kai, M.; Kunert, G.; Gershenzon, J.; D’Auria, J.C. The first step in the biosynthesis of cocaine in Erythroxylum coca: The characterization of arginine and ornithine decarboxylases. Plant Mol. Biol. 2012, 78, 599–615.
- Pegg, A.E. Functions of Polyamines in Mammals. J. Biol. Chem. 2016, 291, 14904–14912.
- Han, L. Studies on Mechanism of Low-Temperature Storage and Polyamine Impact in Cut Flowers of Herbaceous Peony Postharvest Senescence; Shandong Agricultural University: Shandong, China, 2016.
- Ouyang, J.; Song, C.; Chen, D. Research progress on heat-tolerance mechanism and transports of polyamines in plant. Mol. Plant Breed. 2017, 15, 3286–3294.
- De Oliveira, L.F.; Navarro, B.V.; Cerruti, G.V.; Elbl, P.; Minocha, R.; Minocha, S.C.; Dos Santos, A.L.W.; Floh, E.I.S. Polyamine- and Amino Acid-Related Metabolism: The Roles of Arginine and Ornithine are Associated with the Embryogenic Potential. Plant Cell Physiol. 2018, 59, 1084–1098.
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine function in plants: Metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 2019, 9, 1945.
- Hanfrey, C.; Sommer, S.; Mayer, M.J.; Al, E. Arabidopsis polyamine biosynthesis: Absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J. 2010, 27, 551–560.
- Sindhu, R.K.; Cohen, S.S. Subcellular Localization of Spermidine Synthase in the Protoplasts of Chinese Cabbage Leaves. Plant Physiol. 1984, 76, 219–223.
- Hanzawa, Y.; Takahashi, T.; Michael, A.J.; Burtin, D.; Long, D.; Piñeiro, M.; Coupland, G.; Komeda, Y. ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J. 2000, 19, 4248–4256.
- Tavladoraki, P.; Rossi, M.N.; Saccuti, G.; Perez-Amador, M.A.; Polticelli, F.; Angelini, R.; Federico, R. Heterologous Expression and Biochemical Characterization of a Polyamine Oxidase from Arabidopsis Involved in Polyamine Back Conversion. Plant Physiol. 2006, 141, 1519–1532.
- Kim, D.W.; Watanabe, K.; Murayama, C.; Izawa, S.; Niitsu, M.; Michael, A.J.; Berberich, T.; Kusano, T. Polyamine Oxidase5 Regulates Arabidopsis Growth through Thermospermine Oxidase Activity. Plant Physiol. 2014, 165, 1575–1590.
This entry is offline, you can click here to edit this entry!
