Allotetraploid cotton (Gossypium hirsutum and Gossypium barbadense) are cultivated worldwide for its white fiber. Since centuries, conventional breeding approaches increase cotton yield at the cost of extensive erosion of natural genetic variability. Sea Island cotton (G. barbadense) is known for its superior fiber quality, but show poor adaptability as compared to Upland cotton. Hence, there is a dire need to improve the current germplasm resources of Sea Island cotton to develop diverse breeding lines with improved adaptability and excellent agronomic and economic traits. Ethyl methanesulfonate (EMS) is an excellent mutagenic agent that induces genome-wide point mutations to activate the mutagenic potential of plants. In current study, we determined the optimal EMS experimental procedure suitable for construction of cotton mutant library. At M6 generation, mutant library comprised of lines with distinguished phenotypes of the plant architecture, leaf, flower, boll and fiber. Genome wide analysis of SNP distribution and density in yellow leaf mutants reflected the better quality of mutant library. Our mutant collection will serve as the valuable resource for basic research on cotton functional genomics, as well as cotton breeding.
- Ethylmethanesulfonate
- mutant bank
- mutation breeding
- germplasm resources
- Allotetraploid cotton
- plant architecture
- virescent mutants
1. Introduction
Identification of novel alleles in cotton has taken an immense importance in facilitating the genetic improvement and functional genomics research. Conventional breeding approaches have made enormous contributions to the cultivation of cotton varieties; however, increasing biased manipulation of available genetic variability in cotton germplasm resources has resulted in great loss of genetic potential and increased vulnerability to many insect/pests [1]. This selection pressure has reduced the allelic diversity and hampers the efforts to improve the agronomic traits of cotton and limit our understanding about the molecular mechanisms that respond to environmental stresses and pathogens [2]. In this scenario, broadening the genetic base by induced mutation can diversify and create novel changes in the functional genes to develop more germplasm resources [3]. Various mutagenic techniques have been routinely used in cotton gene function studies like T-DNA insertion [4] and gene silencing approaches [5], but these techniques may be impractical, time consuming, and costly in cotton species owing to transgenic barriers. While induced physical and chemical mutagenesis have been demonstrated as an easy and cost-effective way for wide range of genetic mutation. A variety of physical or chemical mutagenic agents have been used to generate genetic variability in cotton, and the most commonly used agents are fast-neutron, gamma-ray bombardments, and ethylmethanesulfonate (EMS). Use of several ionizing radiations and chemical mutagens can be employed to effectively increase the mutation frequency in Gossypium species [6]. Among physical and chemical mutagenic sources, EMS has demonstrated as an excellent mutagenic agent that activates mutagenic potential of crop plants [7]. EMS mutagenesis generates point mutations that are evenly distributed over the whole genome, hence generating large genotypic and phenotypic diversity [1]. EMS effectively replaces the G:C pairing at the nitrogenous base level with any other base pairing through 3-ethyladenine pairing errors bringing 2–10 mutations/Mb of diploid DNA [8]. Mutagenesis in combination with modern genetic tools has the potential to rapidly utilize the genetic variability of targeted loci in cotton for both improving fiber characteristics and abiotic/biotic stress tolerance. EMS-induced mutations in Upland cotton has been used to generate new valuable alleles and traits, leading to development of commercial cultivated varieties [6][9][10][11][12]. Recently, EMS-induced mutant libraries have been constructed in Upland cotton [13] representing beneficial genetic variations. Such collection of mutants represented a broad repertoire of mutants, that may serve as a valuable resource for functional genomic research on complex allotetraploid traits. In EMS-induced mutagenesis, suitable doses of EMS are required to achieve the ideal results. Generally, mutagen doses inducing about 50% lethality (LD50) among M1 plants are considered as appropriate dose because of their high mutation frequency [14]
Sea Island cotton (G. barbadense) is the second most cultivated cotton species, known for its longer staple and superior fiber quality, with a genome size of 2.2 Gb and high content of repetitive elements [15]. After polyploidization, G. barbadense L. evolved to produce a superior fiber quality but show lower diversity than G. hirsutum, which is most cultivated and geographically diverse specie. Only few studies had been conducted on G. barbadense, despite its superiority in fiber quality over G. hirsutum, and accounted for less genetic diversity [16]. Because of its genome complexity, only few genes were identified using the map-based cloning approach. Hence, EMS-based mutagenesis provide reliable source for manipulating genetic mechanism underlying cotton plant architecture, leaf, flower, and fiber related traits is essential for engineering climate-resilient Sea Island cotton varieties. The sympodial and monopodial branches are major determinant of plant architecture and are therefore agronomically important [17]. Leaves are the main source of photo-assimilate in crop plants and photosynthesis is a regulated process that is highly sensitive to environmental stress, as it needs to balance the light energy absorbed by the photosystems with the energy consumed by the metabolic sinks. The chlorophyll (Chl) contents of leaves are positively correlated with photosynthetic efficiency and greatly affect the yield and quality of cotton [18]. Recently, EMS-induced mutation resulted in threonine/isoleucine substitution in a tetratricopeptide repeat-like superfamily protein which is responsible for the short fiber phenotype in Upland cotton [19].
2. Determination of Lethal Dose of EMS Treatment
Genotype ISR was selected for large-scale mutagenic treatment and mutant library construction using the dose and treatment time estimated in preliminary tests. Germination rate of ISR in different concentration of EMS treated for 2 h have been presented in Figure 2. Therefore, for field experiment 2% of EMS dose and 2 h of treatment time was selected as the optimized EMS treatment conditions for cotton.

Figure 1. Germination rate of ISR seeds under different concentrations of EMS.
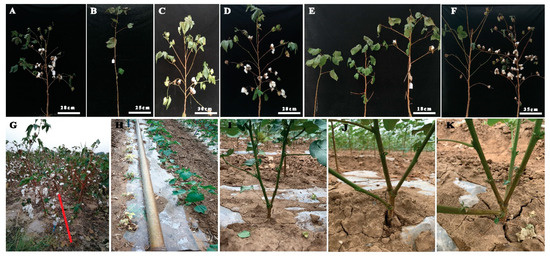
Figure 2. Phenotypic analysis of plant architecture in EMS-induced mutants. (A) Wild type plant architecture, (B) mutant phenotype without fruiting branch, (C) medium height with spreading branches, (D) high yielding mutant as compared to wild type, (E) comparison of fruit branch pattern including without fruiting branch, compact and short fruit branch, longer fruit branches, (F) Longer and spreading branches with different boll bearing capacity, (G) comparison of mutant lines high and low seed cotton yield, (H) comparison of yellow leaf plants with green leaf plants, (I) mutant plant with two main stems, (J) mutant plant with three main stems, (K) mutant plant with four main stems.
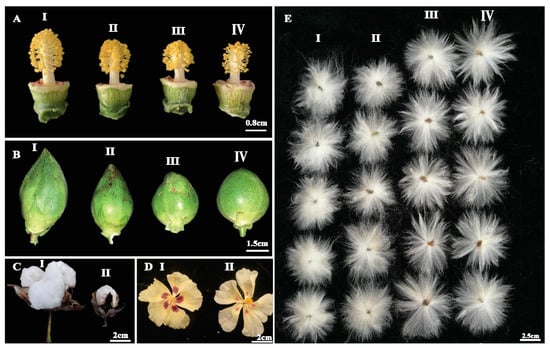
Figure 3. Phenotypic analysis of floral traits in EMS-induced mutants. (A) Comparison of wild type (II) and mutant lines (I, III, and IV) with varying length of flower style and anther numbers, (B) comparison of wild type (II) with mutant lines (I, III, and IV) with different boll size and shape ranging from oblong shape to nearly round boll, (C) evaluation of fully mature cotton boll with big boll size in wild type (I) and small boll size in mutant line (II), (D) comparison of wild type (I) and mutant line (II) for size and intensity of petal spot, (E) comparison of fiber length with I and II as mutants with short fiber length, III represent as fiber length of wild type, IV represent mutant with longer fiber length.
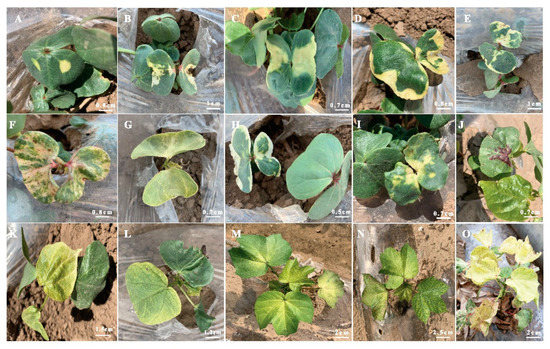
Figure 4. Yellow and yellow-green leaves of EMS-induced mutants (A–J) cotyledon leaves with different shades and positions of yellow color and anthocyanin pigments, (K–O) young true leaves with different shades of yellow and yellow-green color.
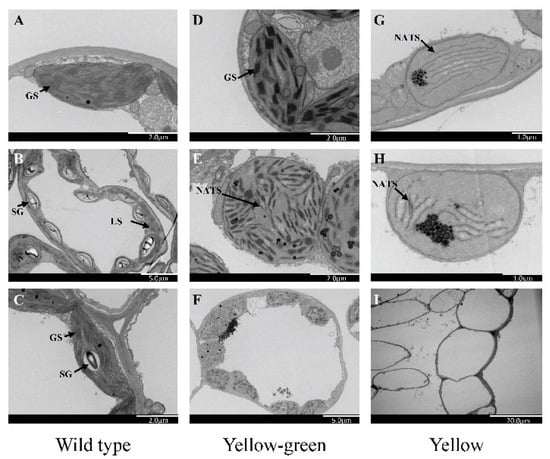
Figure 5. Transmission electron microscopy of the chloroplast ultrastructure, (A–C) Electron micrographs showing mesophyll cells and ultrastructure of chloroplast in wild type, (D–F) yellow-green leaf mutants, (G–I) yellow leaf mutants. GS, granal stack; SG, starch grain; NATS, non-appressed thylakoid stacking; LS, lamellar structure.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21186505
References
- Usman, A.; Cheema, H.M.N.; Sheraz, A.; Khan, I.A.; Waqas, M.; Khan, A.A. COTIP: Cotton TILLING Platform, a Resource for Plant Improvement and Reverse Genetic Studies. Front. Plant Sci. 2016, 7, 1863.
- Zhang, Y.; Wang, X.F.; Li, Z.K.; Zhang, G.Y.; Ma, Z.Y. Assessing genetic diversity of cotton cultivars using genomic and newly developed expressed sequence tag-derived microsatellite markers. GMR 2011, 10, 1462.
- Xu, T.; Bian, N.; Wen, M.; Xiao, J.; Yuan, C.; Cao, A.; Zhang, S.; Wang, X.; Wang, H. Characterization of a common wheat (Triticum aestivum L.) high-tillering dwarf mutant. Theor. Appl. Genet. 2017, 130, 483–494.
- Alonso, J.M. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 1849.
- Wesley, S.V. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001, 27, 581–590.
- Auld, D.L.; Bechere, E.; Ethridge, M.; Becker, W.D.; Cantrell, R.G. Registration of TTU 202-1107-B and TTU 271-2155-C mutant germplasm lines of upland cotton with improved fiber quality. Crop Sci. 2000, 40, 1835–1836
- Aslam, U.; Khan, A.A.; Cheema, H.M.N.; Imtiaz, F.; Malik, W. Kill Curve Analysis and Response of Ethyl Methanesulfonate and γ-rays in Diploid and Tetraploid Cotton. Int. J. Agric. Biol. 2013, 15, 1560–8530.
- Till, B.J.; Cooper, J.; Tai, T.H.; Colowit, P.; Greene, E.A.; Comai, H.L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 2007, 7, 7–19.
- Herring, A.D.; Auld, D.L.; Ethridge, M.D.; Hequet, E.F.; Bechere, E.; Green, C.J.; Cantrell, R.G. Inheritance of fiber quality and lint yield in a chemically mutated population of cotton. Euphytica 2004, 136, 333–339.
- Muthusamy, A.; Jayabalan, N. In vitro induction of mutation in cotton (Gossypium hirsutum L.) and isolation of mutants with improved yield and fiber characters. Acta Physiol. Plant. 2011, 33, 1793–1801.
- Brown, N.; Smith, W.C.; Auld, D.; Hequet, E.F. Improvement of Upland Cotton Fiber Quality through Mutation of TAM 94L-25. Crop Sci. 2013, 53, 452–459.
- Patel, J.D.; Wright, R.J.; Chandnani, R.; Goff, V.H.; Ingles, J.; Paterson, A.H. EMS-mutated cotton populations suggest overlapping genetic control of trichome and lint fiber variation. Euphytica 2016, 208, 597–608.
- Lian, X.; Liu, Y.; Guo, H.; Fan, Y.; Zeng, F. Ethyl methanesulfonate mutant library construction in Gossypium hirsutum L. for allotetraploid functional genomics and germplasm innovation. Plant J. 2020, 103, 858–868.
- Ke, C.; Guan, W.; Bu, S.; Li, X.; Deng, Y.; Wei, Z.; Wu, W.; Zheng, Y.; Lightfoot, D.A. Determination of absorption dose in chemical mutagenesis in plants. PLoS ONE 2019, 14, e0210596.
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748.
- Hinze, L.L.; Elodie, G.; Gore, M.A.; Fang, D.D.; Scheffler, B.E.; Yu, J.Z.; Jones, D.C.; James, F.; Percy, R.G. Genetic diversity of the two commercial tetraploid cotton species in the Gossypium Diversity Reference Set. J. Hered. 2016, 107, 274–286.
- Si, Z.; Liu, H.; Zhu, J.; Chen, J.; Wang, Q.; Lei, F.; Gao, F.; Tian, Y.; Chen, Y.; Chang, L.; et al. Mutation of self-pruning homologs in cotton promotes short-branching plant architecture. J. Exp. Bot. 2018, 69, 2543–2553.
- Liu, J.; Meng, Y.; Lv, F.; Chen, J.; Ma, Y.; Wang, Y.; Chen, B.; Zhang, L.; Zhou, Z. Photosynthetic characteristics of the subtending leaf of cotton boll at different fruiting branch nodes and their relationships with lint yield and fiber quality. Front. Plant Sci. 2015, 6, 747.
- Fang, D.D.; Naoumkina, M.; Thyssen, G.N.; Bechere, E.; Li, P.; Florane, C.B. An EMS-induced mutation in a tetratricopeptide repeat-like superfamily protein gene (Ghir_A12G008870) on chromosome A12 is responsible for the li y short fiber phenotype in cotton. Theor. Appl. Genet. 2020, 133, 271–282.
