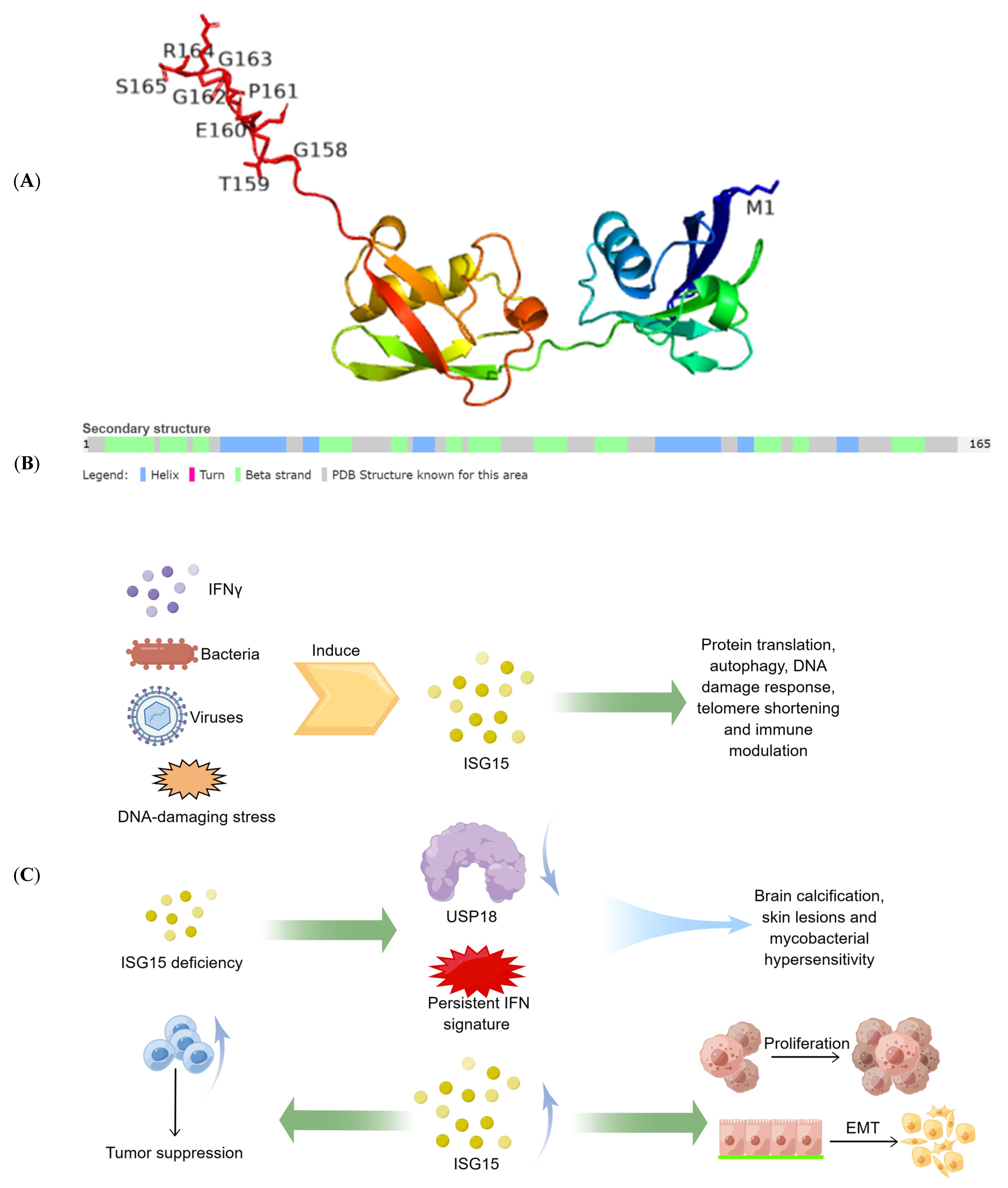
The protein ISG15 encoded by interferon-stimulated gene (ISG) 15 is the first identified member of the ubiquitin-like protein family and exists in the form of monomers and conjugated complexes. Like ubiquitin, ISG15 can mediate an ubiquitin-like modification by covalently modifying other proteins, known as ISGylation. There is growing evidence showing that both the free and conjugated ISG15 are involved in multiple key cellular processes, including autophagy, exosome secretion, DNA repair, immune regulation, and cancer occurrence and progression.
- ISG15
- ISGylation
- cancer
- ubiquitin-like protein
- post-translational modification
1. Introduction
|
UBL Proteins |
E1 |
E2 |
E3 |
Function |
|---|---|---|---|---|
|
UFM1 |
UBA5 |
UFC1 |
UFL1 |
Ufmylation is involved in reticulophagy (also called ER-phagy)-induced in response to endoplasmic reticulum stress [11]. Ufmylation of TRIP4 regulates nuclear receptor-mediated transcription [12]. |
|
SUMO family |
SAE1,SAE2 |
Ubc9 |
PIAS, CBX4 |
Regulating protein stability and the interaction between proteins and subcellular localization [13]. |
|
ISG15 |
UBA7 |
UBCH8 |
HERC5, TRIM25 |
Cancer-promoting or -suppressing; Antiviral infection [14]. |
|
NEDD8 |
NAE1 |
UBE2M, UBE2F |
RBX1, RBX2, FBXO11, c-CBL, DCNL1-5, IAPs, RNF111, RNF111, TFB3, TRIM40 |
Regulating embryonic development, cell division and proliferation [15][16][17]. |
|
UBL3 |
- |
- |
- |
Regulating MHCII and CD86 in human dendritic cells (DCs) and macrophages, regulating immune response [18]. |
|
ATG8 |
ATG7 |
ATG3 |
ATG5-12 |
Regulation of autophagy [19]. |
|
ATG12 |
ATG7 |
ATG10 |
- |
Involving in autophagy vesicle formation [20]. |
|
FAT10 |
UBA6 |
UBE2Z |
Parkin |
Affecting the occurrence, |
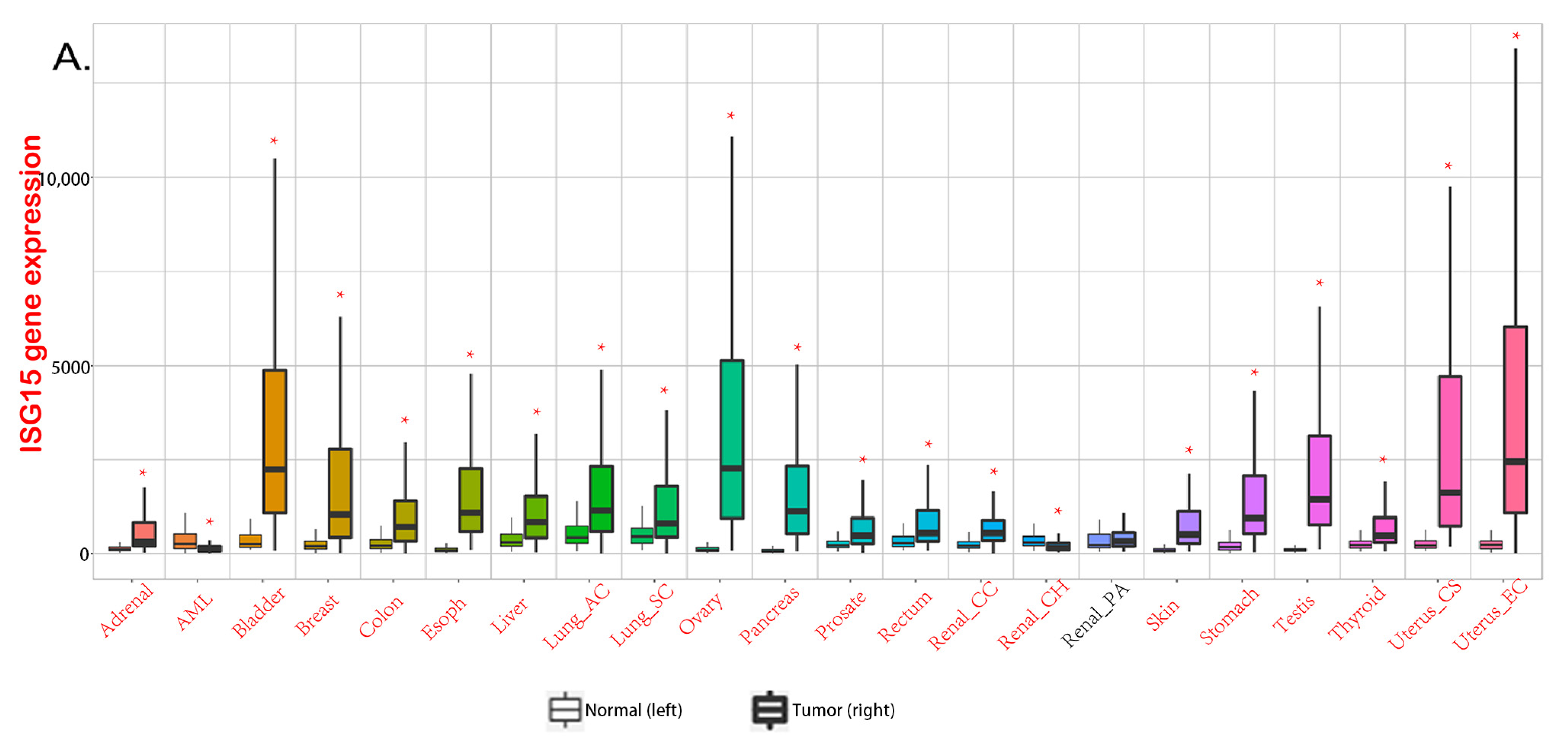
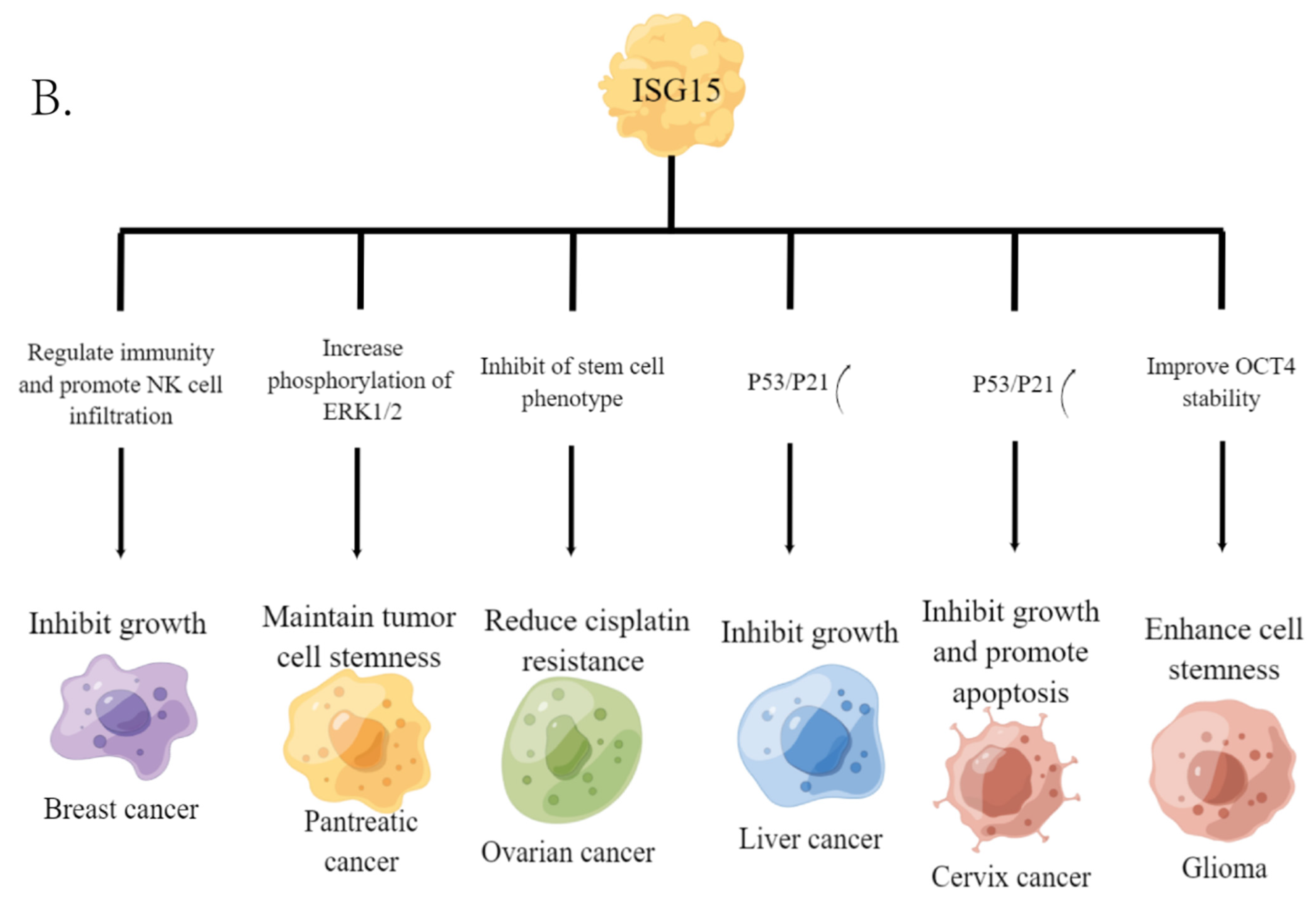
2. Free ISG15 in Cancer
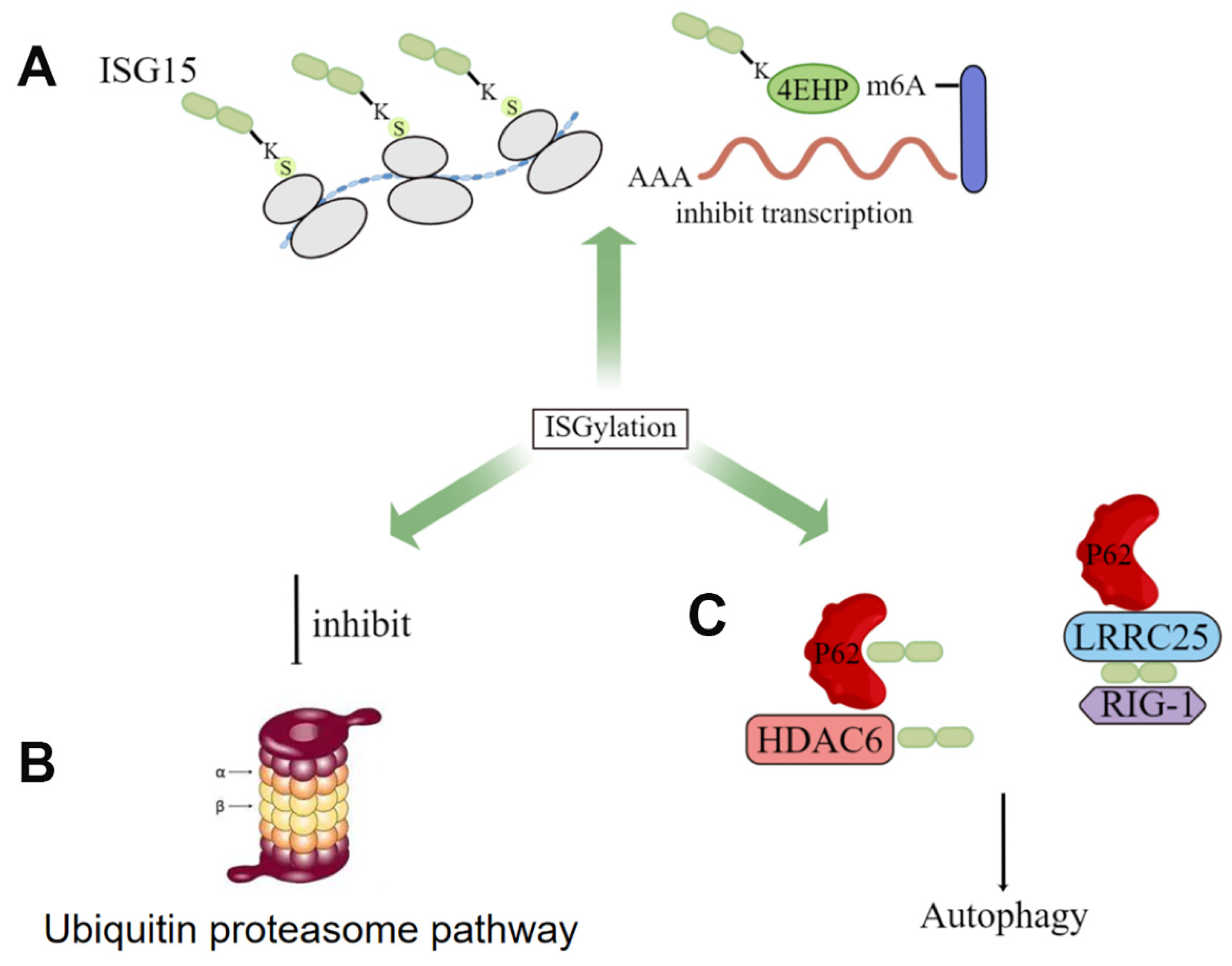
3. ISGylation in Cancer
4. Clinical Applications of ISG15
This entry is adapted from the peer-reviewed paper 10.3390/molecules28031337
References
- Schmid, S.; Hugel, T.; Institute of Physical Chemistry; University of Freiburg; Germany; Signalling research centers BIOSS; Cibss; Albert Ludwigs University. Germany Controlling protein function by fine-tuning conformational flexibility. Elife 2020, 9, e57180.
- Huang, B.; Zhao, Z.; Zhao, Y.; Huang, S. Protein arginine phosphorylation in organisms. Int. J. Biol. Macromol. 2021, 171, 414–422.
- Liu, J.; Guan, D.; Dong, M.; Yang, J.; Wei, H.; Liang, Q.; Song, L.; Xu, L.; Bai, J.; Liu, C.; et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nature 2020, 22, 1056–1063.
- Clarke, S.G. Protein methylation at the surface and buried deep: Thinking outside the histone box. Trends Biochem. Sci. 2013, 38, 243–252.
- Deng, S.; Marmorstein, R. Protein N-Terminal Acetylation: Structural Basis, Mechanism, Versatility, and Regulation. Trends Biochem. Sci. 2020, 46, 15–27.
- Li, N.; Zhang, S.; Xiong, F.; Eizirik, D.L.; Wang, C.-Y. SUMOylation, a multifaceted regulatory mechanism in the pancreatic beta cells. Semin. Cell Dev. Biol. 2020, 103, 51–58.
- Liu, J.; Zhong, L.; Guo, R. The Role of Posttranslational Modification and Mitochondrial Quality Control in Cardiovascular Diseases. Oxidative Med. Cell. Longev. 2021, 2021, 1–15.
- Chen, L.; Liu, S.; Tao, Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target. Ther. 2020, 5, 90.
- Hsu, J.-M.; Li, C.-W.; Lai, Y.-J.; Hung, M.-C. Posttranslational Modifications of PD-L1 and Their Applications in Cancer Therapy. Cancer Res. 2018, 78, 6349–6353.
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577.
- Liang, J.R.; Lingeman, E.; Luong, T.; Ahmed, S.; Muhar, M.; Nguyen, T.; Olzmann, J.A.; Corn, J.E. A Genome-wide ER-phagy Screen Highlights Key Roles of Mitochondrial Metabolism and ER-Resident UFMylation. Cell 2020, 180, 1160–1177.e20.
- Yoo, H.M.; Kang, S.H.; Kim, J.Y.; Lee, J.E.; Seong, M.W.; Lee, S.W.; Ka, S.H.; Sou, Y.-S.; Komatsu, M.; Tanaka, K.; et al. Modification of ASC1 by UFM1 is crucial for ERalpha transactivation and breast cancer development. Mol. Cell 2014, 56, 261–274.
- Pichler, A.; Fatouros, C.; Lee, H.; Eisenhardt, N. SUMO conjugation—A mechanistic view. Biomol. Concepts 2017, 8, 13–36.
- Mirzalieva, O.; Juncker, M.; Schwartzenburg, J.; Desai, S. ISG15 and ISGylation in Human Diseases. Cells 2022, 11, 538.
- Osaka, F.; Kawasaki, H.; Aida, N.; Saeki, M.; Chiba, T.; Kawashima, S.; Tanaka, K.; Kato, S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998, 12, 2263–2268.
- Liakopoulos, D.; Büsgen, T.; Brychzy, A.; Jentsch, S.; Pause, A. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel–Lindau tumor suppressor function. Proc. Natl. Acad. Sci. USA 1999, 96, 5510–5515.
- Xirodimas, D.P.; Saville, M.K.; Bourdon, J.-C.; Hay, R.T.; Lane, D.P. Mdm2-Mediated NEDD8 Conjugation of p53 Inhibits Its Transcriptional Activity. Cell 2004, 118, 83–97.
- Ageta, H.; Tsuchida, K. Post-translational modification and protein sorting to small extracellular vesicles including exo-somes by ubiquitin and UBLs. Cell Mol. Life Sci. 2019, 76, 4829–4848.
- Mijaljica, D.; Prescott, M.; Devenish, R.J. A Late Form of Nucleophagy in Saccharomyces cerevisiae. PLoS ONE 2012, 7, e40013.
- Otomo, C.; Metlagel, Z.; Takaesu, G.; Otomo, T. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013, 20, 59–66.
- Lukasiak, S.; Schiller, C.; Oehlschlaeger, P.; Schmidtke, G.; Krause, P.; Legler, D.F.; Autschbach, F.; Schirmacher, P.; Breuhahn, K.; Groettrup, M. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene 2008, 27, 6068–6074.
- Gong, P.; Canaan, A.; Wang, B.; Leventhal, J.; Snyder, A.; Nair, V.; Cohen, C.D.; Kretzler, M.; D’Agati, V.; Weissman, S.; et al. The ubiquitin-like protein FAT10 mediates NF-kappaB activation. J. Am. Soc. Nephrol. 2010, 21, 316–326.
- Farrell, P.J.; Broeze, R.J.; Lengyel, P. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature 1979, 279, 523–525.
- Haas, A.L.; Ahrens, P.; Bright, P.M.; Ankel, H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 1987, 262, 11315–11323.
- Loeb, K.R.; Haas, A.L. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 1992, 267, 7806–7813.
- Han, H.G.; Moon, H.W.; Jeon, Y.J. ISG15 in cancer: Beyond ubiquitin-like protein. Cancer Lett. 2018, 438, 52–62.
- Tecalco, C.A.; Mejia-Barreto, K. Cell type-dependent regulation of free ISG15 levels and ISGylation. J. Cell Commun. Signal. 2017, 11, 127–135.
- Lowe, J.; McDermott, H.; Loeb, K.; Landon, M.; Haas, A.L.; Mayer, R.J. Immunohistochemical localization of ubiquitin cross-reactive protein in human tissues. J. Pathol. 1995, 177, 163–169.
- Bogunovic, D.; Byun, M.; Durfee, L.A.; Abhyankar, A.; Sanal, O.; Mansouri, D.; Salem, S.; Radovanovic, I.; Grant, A.V.; Adimi, P.; et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science 2012, 337, 1684–1688.
- Chang, Y.-G.; Yan, X.-Z.; Xie, Y.-Y.; Gao, X.-C.; Song, A.-X.; Zhang, D.-E.; Hu, H.-Y. Different Roles for Two Ubiquitin-like Domains of ISG15 in Protein Modification. J. Biol. Chem. 2008, 283, 13370–13377.
- Sadler, A.J.; Williams, B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568.
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622.
- Tecalco-Cruz, A.C.; Cruz-Ramos, E. Protein ISGylation and free ISG15 levels are increased by interferon gamma in breast cancer cells. Biochem. Biophys. Res. Commun. 2018, 499, 973–978.
- Tecalco-Cruz, A.C.; Ramírez-Jarquín, J.O.; Cruz-Ramos, E. Regulation and action of interferon-stimulated gene 15 in breast cancer cells. Hum. Cell 2020, 33, 954–962.
- Tecalco-Cruz, A.C.; Cortés-González, C.C.; Cruz-Ramos, E.; Jarquín, J.O.R.; Romero-Mandujano, A.K.; Sosa-Garrocho, M. Interplay between interferon-stimulated gene 15/ISGylation and interferon gamma signaling in breast cancer cells. Cell Signal. 2019, 54, 91–101.
- Alcalá, S.; Sancho, P.; Martinelli, P.; Navarro, D.; Pedrero, C.; Martín-Hijano, L.; Valle, S.; Earl, J.; Rodríguez-Serrano, M.; Ruiz-Cañas, L.; et al. ISG15 and ISGylation is required for pancreatic cancer stem cell mitophagy and metabolic plasticity. Nat. Commun. 2020, 11, 2682.
- Zhang, Q.; Wang, J.; Qiao, H.; Huyan, L.; Liu, B.; Li, C.; Jiang, J.; Zhao, F.; Wang, H.; Yan, J. ISG15 is downregulated by KLF12 and implicated in maintenance of cancer stem cell-like features in cispla-tin-resistant ovarian cancer. J. Cell Mol. Med. 2021, 25, 4395–4407.
- Wan, X.X.; Chen, H.C.; Khan, M.; Xu, A.H.; Yang, F.L.; Zhang, Y.Y.; Zhang, D.Z. ISG15 inhibits IFN-alpha-resistant liver cancer cell growth. Biomed Res. Int. 2013, 2013, 570909.
- Zhou, M.-J.; Chen, F.-Z.; Chen, H.-C.; Wan, X.-X.; Zhou, X.; Fang, Q.; Zhang, D.-Z. ISG15 inhibits cancer cell growth and promotes apoptosis. Int. J. Mol. Med. 2016, 39, 446–452.
- Dai, Y.; Yu, T.; Yu, C.; Lu, T.; Zhou, L.; Cheng, C.; Ni, H. ISG15 enhances glioma cell stemness by promoting Oct4 protein stability. Environ. Toxicol. 2022, 37, 2133–2142.
- Okumura, F.; Zou, W.; Zhang, D.-E. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 2007, 21, 255–260.
- Okumura, F.; Okumura, A.J.; Uematsu, K.; Hatakeyama, S.; Zhang, D.-E.; Kamura, T. Activation of Double-stranded RNA-activated Protein Kinase (PKR) by Interferon-stimulated Gene 15 (ISG15) Modification Down-regulates Protein Translation. J. Biol. Chem. 2013, 288, 2839–2847.
- Adapala, N.S.; Swarnkar, G.; Arra, M.; Shen, J.; Mbalaviele, G.; Ke, K.; Abu-Amer, Y. Inflammatory osteolysis is regulated by site-specific ISGylation of the scaffold protein NEMO. Elife 2020, 9, e56095.
- Villarreal, D.O.; Wise, M.C.; Siefert, R.J.; Yan, J.; Wood, L.M.; Weiner, D.B. Ubiquitin-like Molecule ISG15 Acts as an Immune Adjuvant to Enhance Antigen-specific CD8 T-cell Tumor Immunity. Mol. Ther. 2015, 23, 1653–1662.
- Desai, S.D.; Haas, A.L.; Wood, L.M.; Tsai, Y.-C.; Pestka, S.; Rubin, E.H.; Saleem, A.; Nur-E-Kamal, A.; Liu, L.F. Elevated Expression of ISG15 in Tumor Cells Interferes with the Ubiquitin/26S Proteasome Pathway. Cancer Res. 2006, 66, 921–928.
- Bhushan, J.; Radke, J.B.; Perng, Y.C.; Mcallaster, M.; Lenschow, D.J.; Virgin, H.W.; Sibley, L.D. ISG15 Connects Autophagy and IFN-γ-Dependent Control of Toxoplasma gondii Infection in Human Cells. mBio 2020, 11, e00852-20.
- Chiok, K.; Pokharel, S.M.; Mohanty, I.; Miller, L.G.; Gao, S.J.; Haas, A.L.; Tran, K.C.; Teng, M.N.; Bose, S. Human Respiratory Syncytial Virus NS2 Protein Induces Autophagy by Modulating Beclin1 Protein Stabilization and ISGylation. mBio 2022, 13, e0352821.
- Yan, W.; Shih, J.H.; Rodriguez-Canales, J.; Tangrea, M.A.; Ylaya, K.; Hipp, J.; Player, A.; Hu, N.; Goldstein, A.M.; Taylor, P.R.; et al. Identification of unique expression signatures and therapeutic targets in esophageal squamous cell carcinoma. BMC Res. Notes 2012, 5, 73.
- Tao, J.; Hua, P.; Wen, J.; Hu, Y.; Yang, H.; Xie, X. Prognostic value of ISG15 mRNA level in drinkers with esophageal squamous cell cancers. Int. J. Clin. Exp. Pathol. 2015, 8, 10975–10984.
- Lin, P.; Yao, Z.; Sun, Y.; Li, W.; Liu, Y.; Liang, K.; Qin, J.; Hou, X.; Chen, L. Deciphering novel biomarkers of lymph node metastasis of thyroid papillary microcarcinoma using proteomic analysis of ultrasound-guided fine-needle aspiration biopsy samples. J. Proteom. 2019, 204, 103414.
- Pitha-Rowe, I.; Hassel, B.; Dmitrovsky, E. Involvement of UBE1L in ISG15 conjugation during retinoid-induced differ-entiation of acute promyelocytic leukemia. J. Biol. Chem. 2004, 279, 18178–18187.
- Weichselbaum, R.R.; Ishwaran, H.; Yoon, T.; Nuyten, D.S.; Baker, S.W.; Khodarev, N.; Su, A.W.; Shaikh, A.Y.; Roach, P.; Kreike, B.; et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chem-otherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 18490–18495.
