Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Candida auris is considered to be an emerging fungal pathogen and is related to high mortality rates, persistent candidemia, inconsistencies in susceptibility testing results and misidentification by available commercial identification systems. Multidrug-resistant (MDR) and pandrug-resistant (PDR) strains are increasingly detected. In Europe, hospital outbreaks caused by C. auris have been reported in the United Kingdom (UK), Italy and Spain.
- Candida auris
- emerging fungal disease
- public health promotion
1. Introduction
From 2009, Candida auris has been considered to be a rising healthcare emergency worldwide. C. auris infections are related to high mortality rates, persistent candidemia, inconsistencies in susceptibility testing results and misidentification by available commercial identification systems. All this must be considered alongside a high risk of treatment failure, which complicates its management [1].
In 2009, C. auris was initially found in Japan [2][3]. However, a retrospective review of the Candida strain found C. auris in South Korea in 1996 [4]. Studies have suggested that C. auris emerged simultaneously and independently in four global regions (South Asia, East Asia, Africa and South America; also named clades I, II, III and IV, respectively). These four clades are genetically distinct [5]. Most recently, a new potential V clade was identified that was isolated from Iran [6]. In the last few years, C. auris infections have increased worldwide [1][7]. In many parts of Africa and Asia, C. auris is now considered to be endemic [8]. In addition, several outbreaks have been reported in European countries such as the United Kingdom (UK), Spain and Italy [1][8][9][10].
Multidrug-resistant (MDR) and pandrug-resistant (PDR) C. auris strains are increasingly detected worldwide. The most frequent resistance is to fluconazole (FLC), followed by amphotericin B (AMB) and voriconazole (VRC). Echinocandin remains the treatment of choice, but resistance can also affect this class of antifungal drugs [1][11].
2. Identification
C. auris was first detected in the external ear canal of a 70-year-old Japanese woman. A 26S ribosomal DNA (rDNA) D1/D2 domain analysis, 18S internal transcribed spacer (ITS) rDNA region sequences and chemotaxonomic studies showed that the newly discovered Candida species (spp.) had a close phylogenetic relationship to the Metschnikowiaceae clade, particularly with C. ruelliae and C. haemulonii [3]. A retrospective study on historical Korean isolates revealed that C. auris strains were initially misidentified as C. haemulonii [12]. A genetic analysis based on ITS 1/2 and D1/D2 sequences showed that C. auris belongs to the Metschnikowiaceae family within the Candida/Clavispora clade such as C. albicans, C. tropicalis, C. haemulonii and C. lusitaniae [3].
The misidentification of C. auris as another yeast species using conventional phenotypic and biochemical methods can be common (Table 1) [2][3]. The thermal tolerance property of growth at temperatures up to 42 °C on CHROMagarTM Candida Plus (CHROMagar, France) has been used to differentiate C. auris from other Candida spp. [13][14]. The diagnosis of C. auris infections includes biochemical-based tests such as analytical profile index strips, VITEK 2, BD Phoenix yeast identification and MicroScan. Nevertheless, these tests lack a comprehensive database for yeast identification [13]. Figure 1 shows C. auris identification.
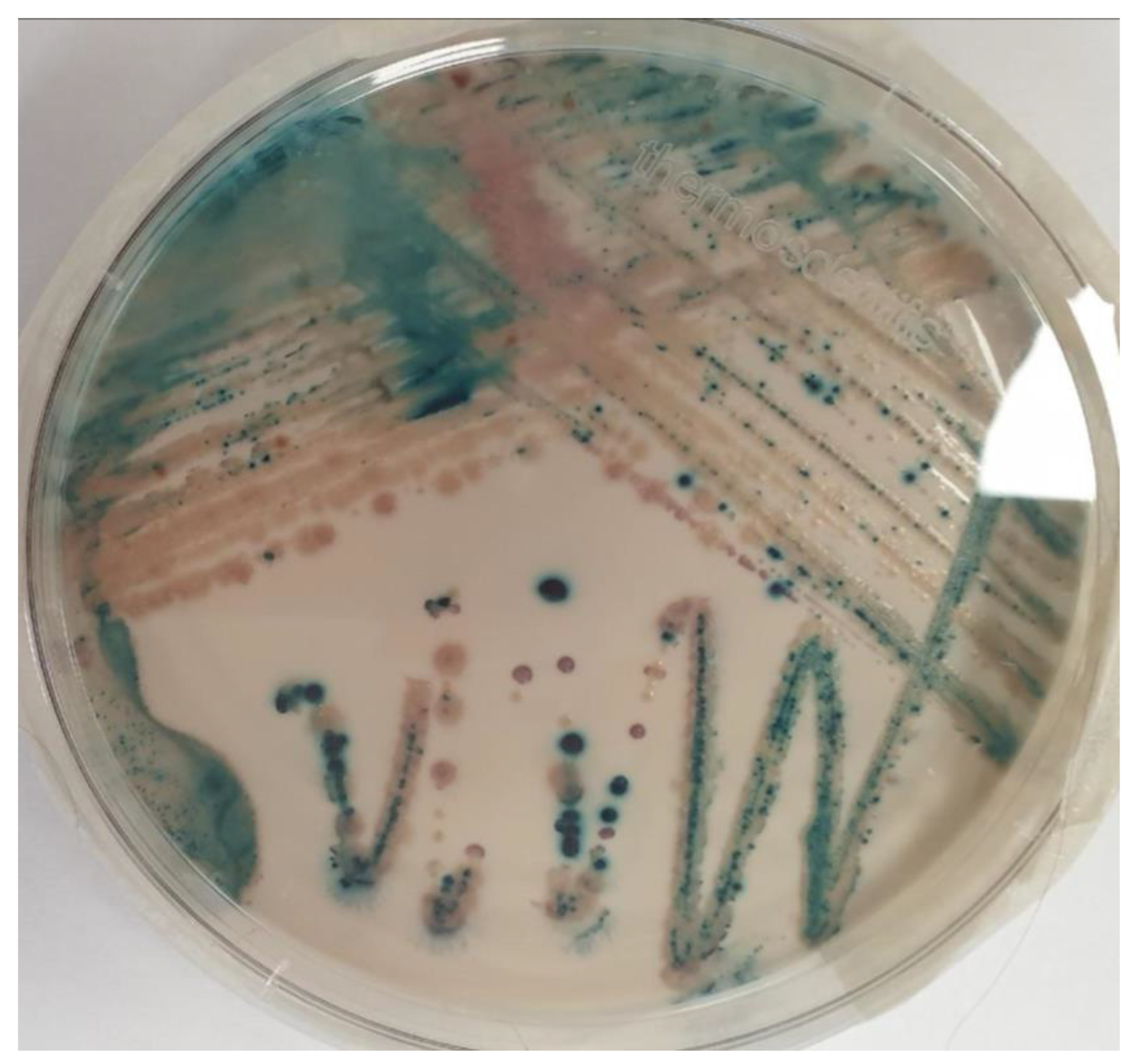
Figure 1. Candida isolates from Brilliance™ Candida Agar Base (Thermo Fisher ScientificTM, Waltham, MA, USA). C. auris (light blue with blue halo colonies), C. krusei (pink and fuzzy colonies) and C. albicans (green–blue colonies).
The identification of yeasts by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analyses has the potential to quickly identify C. auris. However, initial attempts to identify C. auris using this tool were unsuccessful. Following this C. auris isolation across many countries, MALDI-TOF MS added isolates from all four major clades to their FDA-cleared databases [12][13]. In addition, DNA sequencing techniques such as polymerase chain reaction (PCR) have also been used for the identification of C. auris. For example, the PCR amplification of the D1/D2 region and ITS rDNA can be used to differentiate the principal phylogeographic clades of this species, but a further delineation of local hospital clusters required higher resolution methods, including amplified fragment length polymorphism (AFLP) and whole genome sequencing (WGS) analyses [14].
3. Virulence Factors
C. auris can express several virulence factors, including saps and lipases [15]. However, C. auris is less virulent than C. albicans. That characteristic was shown in murine and invertebrate G. mellonella infection models. In murine models, it was demonstrated that C. auris was much more virulent than C. glabrata and C. haemulonii [2][16]. This difference, compared with C. albicans, depended on the inability of C. auris to develop virulence factors such as hyphae or pseudohyphae, which play a critical role in tissue invasion [14]. Furthermore, C. auris is a haploid yeast whereas natural C. albicans isolates are diploid. This could have an essential role in the intrinsically low virulence of C. auris. In FLC-induced haploids, the C. albicans strain reduced their virulence compared with the diploid form [2][17]. The filamentous cells of C. auris are poorly implicated in its virulence during systemic infections, but could play a role in skin and environmental surface colonization [2].
4. Antifungal Resistance
FLC and echinocandins are the most used antifungal drugs to treat candidemia. Unfortunately, FLC (or other azole) resistance is common. A recent meta-analysis from Sekyere et al. showed that the most frequent resistance was to FLC (44.29%), followed by AMB (15.46%), VRC (12.67%), caspofungin (CAS) (3.48%), flucytosine (FC) (1.95%), itraconazole (ITZ) (1.81%), isavuconazole (ISA) (1.53%), posaconazole (POS) (1.39%), anidulafungin (AFG) (1.25%) and micafungin (MFG) (1.25%) [11][12]. MDR C. auris strains have been reported in several cases, showing resistance phenotypes to FLC and AMB [18]. Resistance to echinocandins is not so frequent. Chen et al. found that the resistance rates to CAS, MFG and AFG were 12.1%, 0.8% and 1.1%, respectively. However, almost all isolates resistant to CAS were from India (23.6%) [19].
The molecular mechanism for azole resistance in C. auris is mainly related to alterations in the lanosterol demethylase enzyme, which is encoded by the ERG11 gene. C. auris can also encode ATP-binding cassette (ABC) and major facilitator superfamily (MFS) efflux pumps, which are essential mechanisms of antifungal resistance, especially during the initial stages of biofilm development. When resistance to echinocandins; occur, it is due to mutations in FKS genes that encode a subunit of the β-D-glucan synthase. Moreover, changes to the cell membrane sterol and/or a given point mutation are potential mechanisms of AMB resistance [13][20].
Unfortunately, no antifungal susceptibility breakpoints for C. auris are currently standardized for the Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST). Therefore, the Centers for Disease Control and Prevention (CDC) defined a C. auris-specific antifungal susceptibility interpretation based on a close phylogenetic relationship to other Candida spp. The correlation between the microbiologic breakpoints and clinical outcomes is not known. The current breakpoints are summarized in Table 1 [21].
Table 1. C. auris-specific antifungal susceptibility interpretation according to CDC [21].
| Antifungal | MIC | Interpretation |
|---|---|---|
| Fluconazole | ≥32 | Isolates with MIC ≥ 32 were shown to have a mutation of the Erg11 gene |
| Voriconazole | NA | Consider using fluconazole susceptibility as a surrogate for other azoles. Occasionally, isolates that are resistant to fluconazole may respond to voriconazole |
| Amphotericin B | ≥2 | Isolates with a MIC of ≥2 should be considered to be resistant |
| Anidulafungin | ≥4 | Breakpoints are based on the distribution of echinocandin MICs of approximately 100 isolates from diverse geographic locations |
| Caspofungin | ≥2 | |
| Voriconazole | ≥4 |
5. Risk Factors and Mortality Rates
Most C. auris cases have escalated within the last few years. The reported isolates were mainly isolated in males (64.76%). No reason has been given for the C. auris distribution by gender. Local variables and the health diversity of countries could play a role in the increase in C. auris male case rates. Patients with C. auris infections frequently presented several other underlying health comorbidities such as diabetes, sepsis, pulmonary diseases, bacterial pneumonia, renal diseases, transplants, immunosuppression, solid tumors, cardiovascular diseases, chronic otitis media and liver diseases [1].
The risk factors for C. auris infections are similar to other Candida spp. generic risk factors. Most frequently, infections occur in hospitalized patients, especially those admitted to the intensive care unit (ICU) or those who underwent surgery in the previous 30 days. Moreover, central venous catheters, hemodialysis catheters and permanent urinary catheters could be related to invasive C. auris infections [1][20][22].
Even with an appropriate antifungal treatment, invasive candidiasis has a mortality rate of up to 30–40%. Currently, there is limited information on specific C. auris-case fatality rates. However, several authors have suggested that the mortality rate of invasive C. auris infections is comparatively higher than that of Candida spp. For C. auris, the crude mortality rate was estimated to be 30% to 72% [1][18][23][24].
This entry is adapted from the peer-reviewed paper 10.3390/healthcare11030425
References
- Sekyere, J.O. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. MicrobiologyOpen 2018, 7, e00578.
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921.
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44.
- Kwon, Y.J.; Shin, J.H.; Byun, S.A.; Choi, M.J.; Won, E.J.; Lee, D.; Lee, S.Y.; Chun, S.; Lee, J.H.; Choi, H.J.; et al. Candida auris Clinical Isolates from South Korea: Identification, Antifungal Susceptibility, and Genotyping. J. Clin. Microbiol. 2019, 57, e01624-18.
- Chow, N.A.; Muñoz, J.F.; Gade, L.; Berkow, E.L.; Li, X.; Welsh, R.M.; Forsberg, K.; Lockhart, S.R.; Adam, R.; Alanio, A.; et al. Tracing the Evolutionary History and Global Expansion of Candida auris Using Population Genomic Analyses. MBio 2020, 11, e03364-19.
- Spruijtenburg, B.; Badali, H.; Abastabar, M.; Mirhendi, H.; Khodavaisy, S.; Sharifisooraki, J.; Armaki, M.T.; de Groot, T.; Meis, J.F. Confirmation of fifth Candida auris clade by whole genome sequencing. Emerg. Microbes Infect. 2022, 11, 2405–2411.
- Lane, C.R.; Seemann, T.; Worth, L.J.; Easton, M.; Pitchers, W.; Wong, J.; Cameron, D.; Azzato, F.; Bartolo, R.; Mateevici, C.; et al. Incursions of Candida auris into Australia, 2018. Emerg. Infect. Dis. 2020, 26, 1326–1328.
- Hinrichs, C.; Wiese-Posselt, M.; Graf, B.; Geffers, C.; Weikert, B.; Enghard, P.; Aldejohann, A.; Schrauder, A.; Knaust, A.; Eckardt, K.; et al. Successful control of Candida auris transmission in a German COVID-19 intensive care unit. Mycoses 2022, 65, 643–649.
- Di Pilato, V.; Codda, G.; Ball, L.; Giacobbe, D.R.; Willison, E.; Mikulska, M.; Magnasco, L.; Syamaladevi, R.M.; Tang, J.; Villa-Rojas, R.; et al. Molecular Epidemiological Investigation of a Nosocomial Cluster of C. Auris: Evidence of Recent Emergence in Italy and Ease of Transmission during the COVID-19 Pandemic. J. Fungi. 2021, 7, 140.
- Kohlenberg, A.; Struelens, M.J.; Monnet, D.L.; Plachouras, D.; The Candida auris survey collaborative group. Candida auris: Epidemiological situation, laboratory capacity and preparedness in European Union and European Economic Area countries, 2013 to 2017. Eurosurveillance 2018, 23, 18–00136.
- Ademe, M.; Girma, F. Candida auris: From Multidrug Resistance to Pan-Resistant Strains. Infect. Drug Resist. 2020, 13, 1287–1294.
- Oh, B.J.; Shin, J.H.; Kim, M.-N.; Sung, H.; Lee, K.; Joo, M.Y.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Biofilm Formation and Genotyping of Candida haemulonii, Candida pseudohaemulonii, and a Proposed New Species (Candida auris) Isolates from Korea. Med. Mycol. 2011, 49, 98–102.
- Spivak, E.S.; Hanson, K.E. Candida auris: An Emerging Fungal Pathogen. J. Clin. Microbiol. 2018, 56, e01588-17.
- Lockhart, S.R.; Lyman, M.M.; Sexton, D.J. Tools for Detecting a “Superbug”: Updates on Candida auris Testing. J. Clin. Microbiol. 2022, 60, e0080821.
- Yue, H.; Bing, J.; Zheng, Q.; Zhang, Y.; Hu, T.; Du, H.; Wang, H.; Huang, G. Filamentation in Candida auris, an emerging fungal pathogen of humans: Passage through the mammalian body induces a heritable phenotypic switch. Emerg. Microbes Infect. 2018, 7, 188.
- Fakhim, H.; Vaezi, A.; Dannaoui, E.; Chowdhary, A.; Nasiry, D.; Faeli, L.; Meis, J.F.; Badali, H. Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses 2018, 61, 377–382.
- Hickman, M.A.; Zeng, G.; Forche, A.; Hirakawa, M.P.; Abbey, D.; Harrison, B.D.; Wang, Y.-M.; Su, C.-H.; Bennett, R.J.; Wang, Y.; et al. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 2013, 494, 55–59.
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2017, 64, 134–140.
- Chen, J.; Tian, S.; Han, X.; Chu, Y.; Wang, Q.; Zhou, B.; Shang, H. Is the superbug fungus really so scary? A systematic review and meta-analysis of global epidemiology and mortality of Candida auris. BMC Infect. Dis. 2020, 20, 827.
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Abbasi, A.F.; Prakash, S.; Mangat, J.; Hosein, Z.; Haider, N.; Chan, J. Candida auris: An Overview of the Emerging Drug-Resistant Fungal Infection. Infect. Chemother. 2022, 54, 236–246.
- Antifungal Susceptibility Testing and Interpretation|Candida Auris|Fungal Diseases|CDC. Available online: https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html (accessed on 6 October 2022).
- de Cássia Orlandi Sardi, J.; Silva, D.R.; Mendes-Giannini, M.J.S.; Rosalen, P.L. Candida auris: Epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb. Pathog. 2018, 125, 116–121.
- Sikora, A.; Zahra, F. Candida Auris. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: http://www.ncbi.nlm.nih.gov/books/NBK563297/ (accessed on 25 January 2023).
- Chakrabarti, A.; Sood, P.; Rudramurthy, S.M.; Chen, S.; Kaur, H.; Capoor, M.; Chhina, D.; Rao, R.; Eshwara, V.K.; Xess, I.; et al. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensiv. Care Med. 2015, 41, 285–295.
This entry is offline, you can click here to edit this entry!
