Microbiome is the preeminent factor to maintain the efficacy of vaccine platform. The inception of herd immunity in society is depending upon food habits, microbiome symbiosis, environmental factors, and network among people with each other. Rigorous pan-India polio vaccination program for the last 30 years develops heterologous immunity providing cardinal protection against the COVID-19.
- SARS-CoV-2, herd immunity, polio virus
1. Introduction
Herd immunity, or community immunity, is fundamentally a concept of acquiring immunity through natural infection, or mass vaccination, in a particular population. While the purpose of individual vaccination is to prevent or reduce the chances of recurrent infection, in public health, the goal of herd immunity is to increase immunization efficacy to control or eradicate the infection in a particular cohort. Herd immunity is evidently a sequential process, because natural or vaccine-based immunity is lost over time through the waning of immune individuals due to death, and arrival of newly susceptible individuals due to birth or migration. Thus, to sustain herd immunity, it is important to vaccinate at regular intervals[1]. Herd immunity recuperates the protection against pathogens that are contagious in nature [2][3][4]. The successful eradication of several viral diseases such as smallpox [5] and poliovirus, reduction in transmission of pertussis and protection against influenza, pneumococcal disease, cholera [6] and rotavirus[7]was possible only through expanded immunization programs that have been fundamental factors for evolving herd immunity[8]. In addition, the level of vaccination needed to be achieved through herd immunity varies depending upon the frequencies of secondary infections [3][9]. For example, in measles, a highly contagious viral disease, one person can infect up to 18 individuals. Thus, 95% of the people are required to be immune in order to achieve herd immunity. The new SARS-CoV-2 has a lower infection rate than measles. On average, each infected person can pass the virus to two or three new people [10]. This means that herd immunity should be achieved when around 60% of a particular population becomes exposed to coronavirus disease 2019 (COVID-19) [11].
Here, we outline several factors that can directly or indirectly alter the acquisition of immunity. Among those factors, the gut microbiota is an essential element that contributes to shaping individual immunity and thereby mapping the population immunity. Gut microbiota, along with nutrients and environmental factors, play an indispensable and cumulative role in establishing herd immunity against any infectious disease. We propose that heterologous immunity is the most important factor contributing protection against SARS-CoV-2 infection in India. The robust poliovirus vaccination program developed herd immunity. This same method has the ability to deliver the protection against coronavirus infection.
2. Putting the Microbiome, the Second Brain to Develop the Immune System
To generate efficient herd immunity, the maturation of the immune system is an essential and fundamental factor. Alterations of the microbiota by any means could pose an negative impact on the immune system’s development which upsets the vaccination program and subsequently, herd immunity [12][13]. Collectively, the microbial community residing in our body is termed as the microbiota. Microbiota, the ‘second brain’ of the body, outnumber the total cells of the human body by several fold. Ninety-nine percent of the total genome of human comes from microbiota. One percent comes from 23,000 genes of the body’s own cells [14][15][16]. The gut microbiota dysbiosis significantly affects the development and function of both innate and adaptive immunity [17][18]. The human microbiome regulates several functions of the body such as nutrient metabolism, intestinal barrier functions, shaping of the immune system, etc. It supports the prevention of several diseases and contributes to improving the genetic diversity in the population [19]. The absence of normal microbiota aggravates the maturation of the immune system, affecting both structure and functions as suggested by studies in germ-free (GF) mice[20][21]. However, by restoring microbiota symbiosis, all these effects can be reinstated.
In addition, the microbiome seems to provide a plausible explanation about the differential response of vaccination at both individual and population level. Gut microbiota have specific immunomodulatory properties [22]. Bacteroides fragilis, a Gram-negative bacteria (Phylum-Bacteroidetes) regulates mucosal tolerance to self-antigens by maintaining T-cell homeostasis, preventing T-helper-1 (Th1)/Th17 balance [23] and induces suppressive forkhead box p3+ (Foxp3+) T-regulatory cells (Treg) functions by encouraging anti-inflammatory cytokines such as interleukin-10 (IL-10) and transforming growth factor-β (TGF-β) [24]. Similar preferment of Treg differentiation has been shown with the presence of Clostridia sp. in the colon, but not in the small intestine in maintaining the immune cell homeostasis [25]. Additionally, differentiation of Th17 cells and mucosal immunoglobulin-A (IgA) secretion are possible only with the colonization of segmented filamentous bacteria (SFB) [26]. Several mucosal pathogens such as bacteria, virus and fungi uphold both Th17 and IgA. The ecology of the microbiome is crucial, attaining the highest efficacy of vaccination since infants born because the immunization process has mostly been accomplished during childhood (Figure 1). An infant’s microbiome is determined by the maternal–offspring exchange of microbiota[27]. It becomes similar to the adult microbiome by three years of age. Until then, it is highly variable[28] and grows uninterrupted, along with the development of the immune system[20]. The total microbiome composition of infants is mostly occupied by the phyla such as Firmicutes, Tenericutes, Proteobacteria, Bacteroidetes and Fusobacteria based on commensal microbiota harbors in the placenta [29][30][31]. The variation of commensals is also related to mode of infant delivery. The intestinal flora of neonates born by vaginal delivery resemble the maternal vaginal flora. They are prevalent with Lactobacillus sp. and Prevotella sp. [32]. In another report, vaginal delivery supports higher existence of facultative anaerobic organisms like Escherichia coli, Staphylococcus sp., Streptococcus sp. and other Enterobacteriaceae (Phylum Proteobacteria) for first few days of birth[27][33]. Nonetheless, caesarean babies are predominantly colonized with maternal-skin microbiota such as Clostridium, Staphylococcus, Propionibacterium and Corynebacterium [34][35]. After birth, milk feeding has the most significant impact on changing the microbiome composition of children. Breast milk provides increased prevalence of Bifidobacterium sp. (phylum Actinobacteria) in the intestinal track of infants. Infants are also occupied with higher abundance of Staphylococcus sp. Streptococcus sp., Lactobacillus sp. (phylum Firmicutes), Serratia sp., Ralstonia sp. (phylum Proteobacteria), Corynebacterium sp. (phylum Actinobacteria), etc. in their gut[36]. In contrast, there is a richness of aerobic bacteria and lower frequencies of Bifidobacterium sp. in the guts of bottle milk-fed infants[37][38][39]. The neonatal gut microbiota not only programs the metabolic function, but also educates the naïve immune system, without which, the success of vaccination will be precluded.
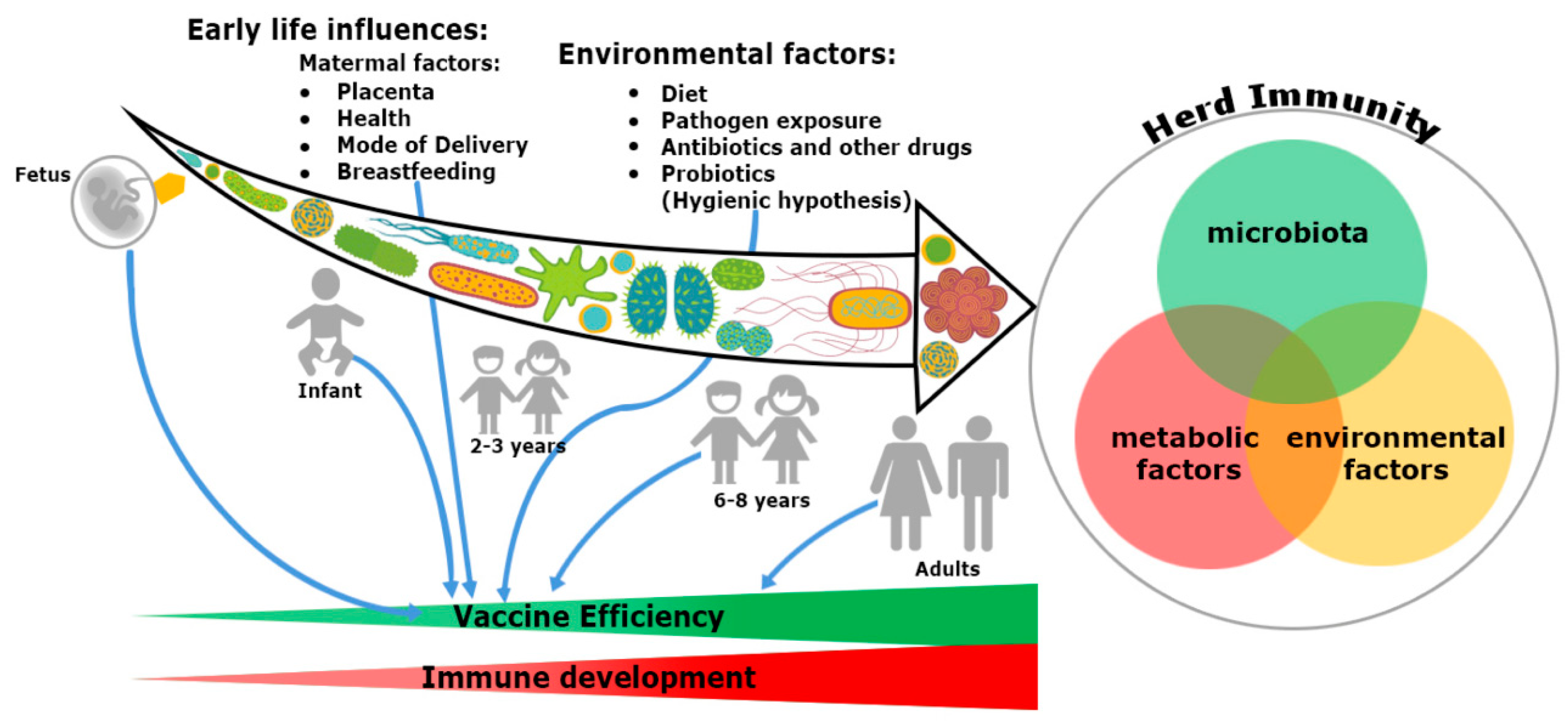
Figure 1. Trinity of the immune system development and vaccine efficiency. The immune system and microbiota mutually co-evolve together in a symbiotic relationship. The impact of microbiome on the immune system development cannot be ignored. From fetus to adulthood, microbiomes are synchronized by maternal transfer and environmental factors. Early maternal factors such as mode of delivery, breastfeeding, antibiotics and diets all influence the immune system. Hence, all have an impact on subsequent immunological responses to many vaccines. Development of herd immunity in a community against any infection is the result of a complex outcome of host-specific factors such as microbiota, metabolism and environmental conditions. Malnutrition and repeated gastrointestinal infections reduce many vaccines’ efficacy. Microbial dysbiosis, along with environmental enteropathy (EE) influences undernourishment. It impairs the immune system development and decreases the efficiency of vaccines in a community, thereby compromising herd immunity.
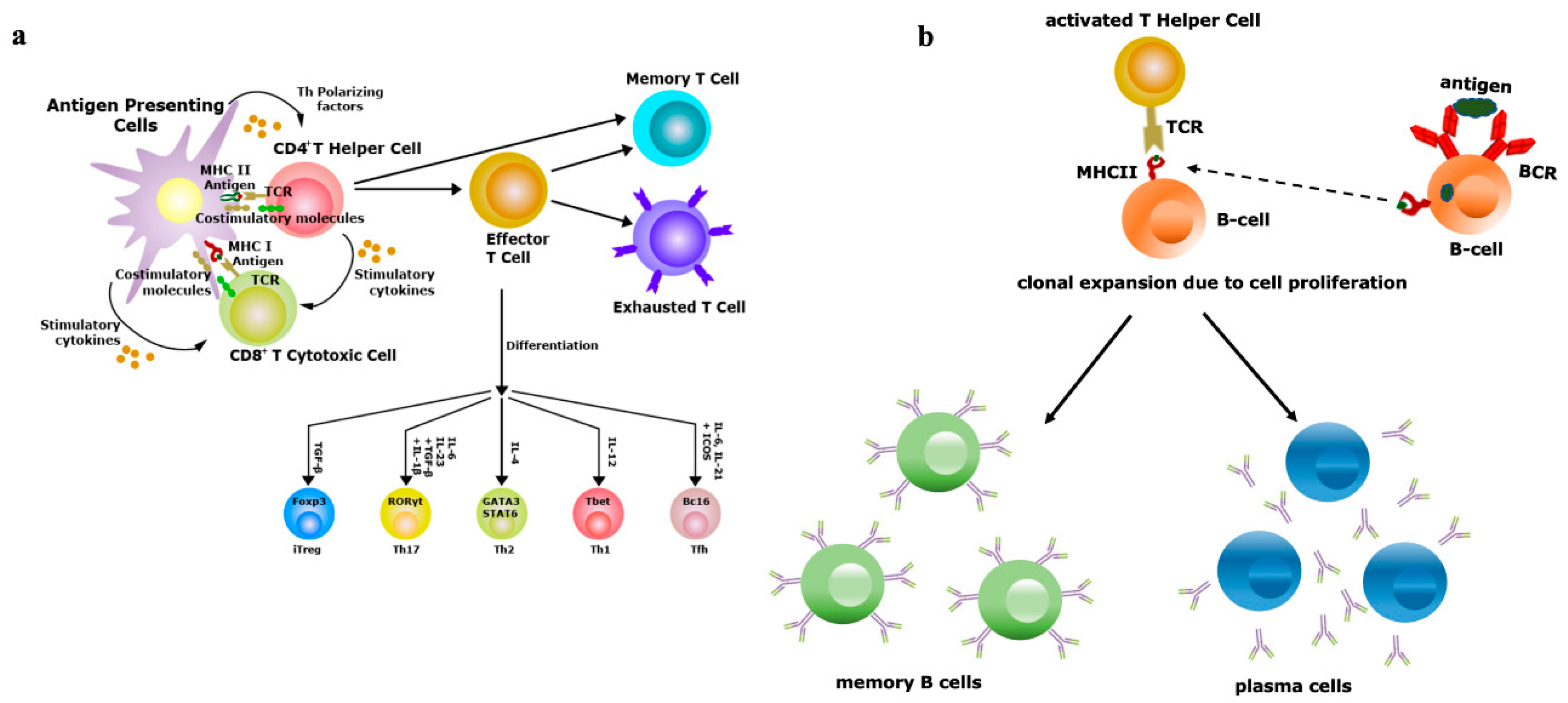
Figure 2. Vaccine efficiency depends on the repertoire diversity of T-cells and B-cells. (a) Upon contact with antigen-presenting cells (APCs) through the T-cell receptor (TCR)–antigen–major histocompatibility complex (MHC) complex, naïve T-cells are differentiated into effector and memory T-cells. Based on the antigenic property, effector T-cells are of two types, i.e., CD4+ T-helper (Th) or CD8+ T-cytotoxic (Tc) cells. Antigens exposed by MHCII binds with TCRs of Th cells and antigens exposed by MHCI binds with TCRs of Tc cells. Th cells have the potential to further differentiate into Th1, Th2, Th17 and induced regulatory T-cells (iTreg), a process controlled by the lineage-specific transcription factors and effector cytokines produced by APCs. Effectiveness of memory T-cells decides the efficacy of vaccination; (b) B-cells (APCs) recognize antigens by their B-cell receptors (BCRs), followed by internalization of antigens and presented to Th cells, which are specific to same antigen. The TCRs of Th cells interact with antigens exposed by MHCII of B-cells. Then, activated B-cells trigger their own proliferation and differentiate into antibody-secreting plasma cells and memory B-cells. Activation and class switching of B-cells is the key factor for the success of immunization.
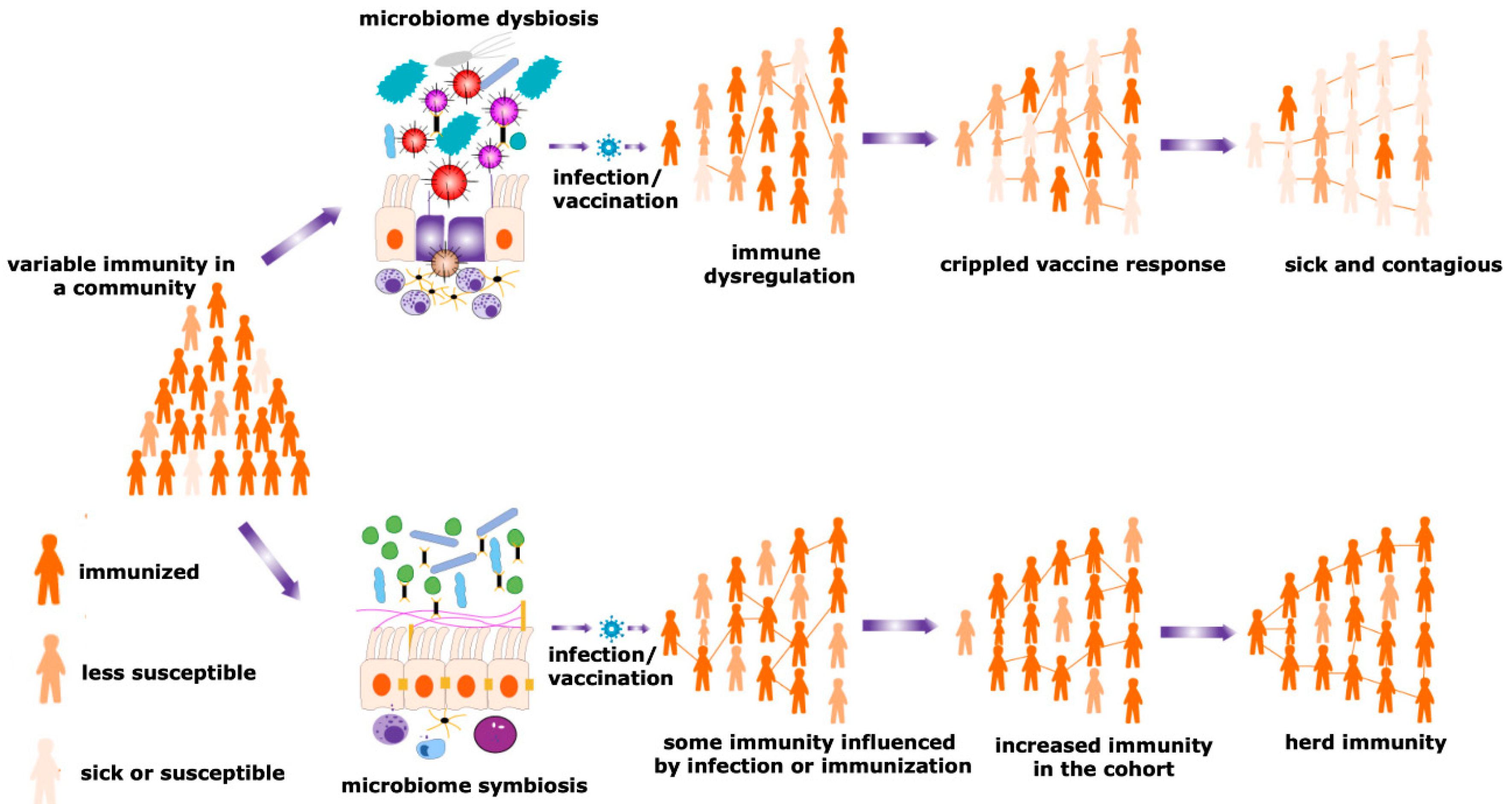
Figure 3. Herd immunity and microbiota. Maturity of the herd immunity against any infectious disease is a significant means of acquiring protection during an epidemic or pandemic situations. A spatial community is occupied by people having a variable degree of resistance against a particular infection. Microbial symbiosis regulates pathogen transmission dynamics and the efficacy of vaccination among different individuals in a population, promoting the development of herd immunity. In contrast, dysbiosis of the microbiome causes immune dysregulation, generating the suboptimal immune response and negative impacts on a vaccination program. Therefore, the number of sick individuals increases in a particular cohort. This reduces the chances of evolving herd immunity and increases the sick and contagious patients in a community.
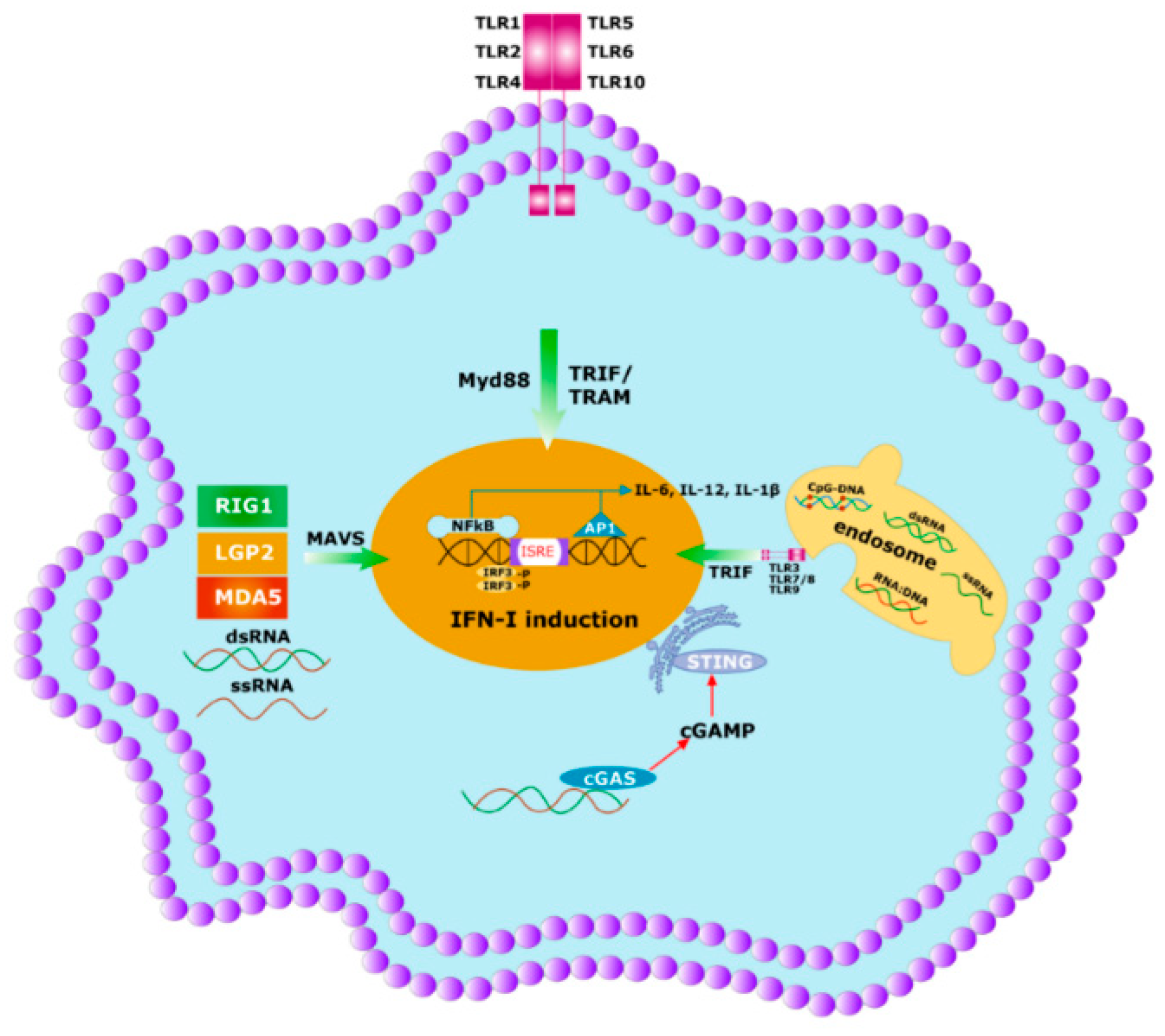
Figure 4. Initial immune response to any pathogen involves the activation of innate immunity. Innate immunity is activated by germline encoded PRRs which recognize PAMPs and DAMPs of the microbes. Specific PRRs mainly include membrane-bound several TLRs, cytosolic DNA sensor, e.g., STING, Rig -1 like receptor (RLR), MDA5, NOD-like receptor, all of which coordinate with the host innate immune responses through the activation of the nuclear factor κB (NF-κB), activator protein 1 (AP-1) and interferon regulatory factor (IRF)-signaling pathway which triggers the antiviral interferons type I (IFNI) pathway. Activated innate immunity can commence the adaptive immunity by triggering both the T-cells and B-cells.
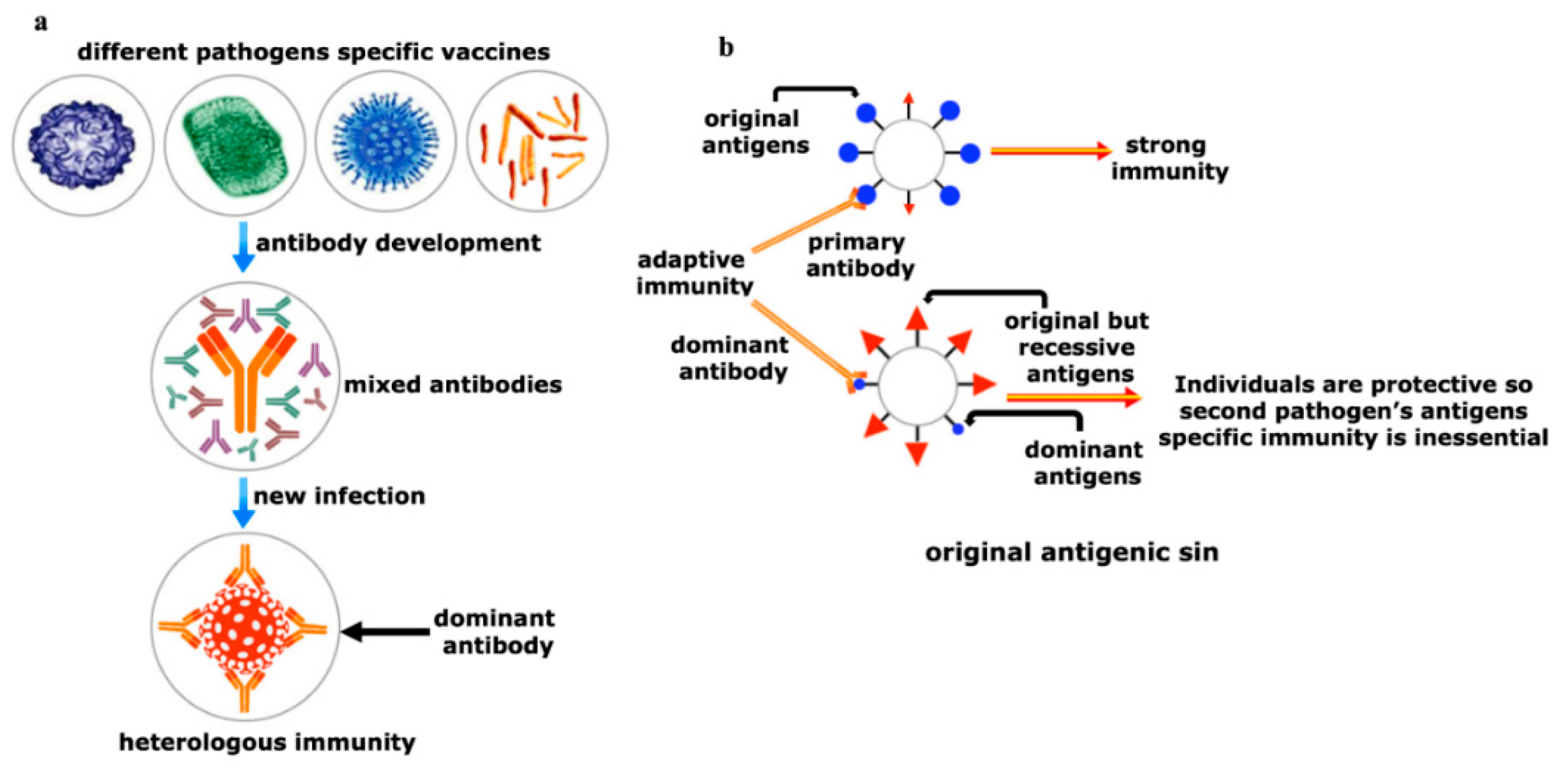
Figure 5. Yin and Yang of the heterologous immunity. (a) Heterologous immunity develops from the opinion where the antibody of one pathogen may protect an individual from a similar or in some cases even from phylogenetically distinct invaders. Vigorous immunization program with several vaccine candidates is required to improve protective response against any new incoming pathogen having comparable antigens. Both humoral and cellular cross-reactive heterologous memory responses may influence the foundation of the natural course of herd immunity against any new infection; (b) original antigenic sin refers to a phenomenon where the development of immunity against pathogens or antigens is shaped by the first exposure of related pathogens. The memory antibodies generated as a result of first exposure will interact with similar antigens of second pathogen for neutralizing it. This anamnestic recall against the second pathogen dampens the probability of immune system activation against original antigens of the second pathogen. This phenomenon is reasonably applicable to SARS-CoV-2 infection where its infection rate is very mild.
This entry is adapted from the peer-reviewed paper 10.3390/v12101150
References
- Nokes, D.J.; Anderson, R.M. The use of mathematical models in the epidemiological study of infectious diseases and in the design of mass immunization programmes. Epidemiol. Infect. 1988, 101, 1–20.
- Anderson, R.M.; May, R.M. Vaccination and herd immunity to infectious diseases. Nature 1985, 318, 323–329.
- Fine, P.; Eames, K.; Heymann, D.L. “Herd immunity”: A rough guide. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2011, 52, 911–916.
- John, T.J.; Samuel, R. Herd immunity and herd effect: New insights and definitions. Eur. J. Epidemiol. 2000, 16, 601–606.
- Lane, J.M. Mass vaccination and surveillance/containment in the eradication of smallpox. Curr. Top. Microbiol. Immunol. 2006, 304, 17–29.
- Ali, M.; Emch, M.; von Seidlein, L.; Yunus, M.; Sack, D.A.; Rao, M.; Holmgren, J.; Clemens, J.D. Herd immunity conferred by killed oral cholera vaccines in Bangladesh: A reanalysis. Lancet 2005, 366, 44–49.
- Mast, T.C.; Wang, F.T.; Su, S.; Seeger, J.D. Evidence of herd immunity and sustained impact of rotavirus vaccination on the reduction of rotavirus-related medical encounters among infants from 2006 through 2011 in the United States. Pediatr. Infect. Dis. J. 2015, 34, 615–620.
- Kim, T.H.; Johnstone, J.; Loeb, M. Vaccine herd effect. Scand. J. Infect. Dis. 2011, 43, 683–689.
- Metcalf, C.J.E.; Ferrari, M.; Graham, A.L.; Grenfell, B.T. Understanding herd immunity. Trends Immunol. 2015, 36, 753–755.
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 27.
- Kwok, K.O.; Lai, F.; Wei, W.I.; Wong, S.Y.S.; Tang, J.W.T. Herd immunity—Estimating the level required to halt the COVID-19 epidemics in affected countries. J. Infect. 2020, 80, e32–e33.
- Ferreira, R.B.; Antunes, L.C.; Finlay, B.B. Should the human microbiome be considered when developing vaccines? PLoS Pathog. 2010, 6, e1001190.
- Lynn, D.J.; Pulendran, B. The potential of the microbiota to influence vaccine responses. J. Leukoc. Biol. 2018, 103, 225–231.
- Guttinger, S. The anti-vaccination debate and the microbiome: How paradigm shifts in the life sciences create new challenges for the vaccination debate. EMBO Rep. 2019, 20.
- Lederberg, J.; McCray, A.T. Ome SweetOmics—A genealogical treasury of words. Scientist 2001, 15, 8.
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848.
- Cerf-Bensussan, N.; Gaboriau-Routhiau, V. The immune system and the gut microbiota: Friends or foes? Nat. Rev. Immunol. 2010, 10, 735–744.
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14.
- Grice, E.A.; Segre, J.A. The human microbiome: Our second genome. Annu. Rev. Genom. Hum. Genet. 2012, 13, 151–170.
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544.
- Magwira, C.A.; Taylor, M.B. Composition of gut microbiota and its influence on the immunogenicity of oral rotavirus vaccines. Vaccine 2018, 36, 3427–3433.
- Ivanov, I.I.; Honda, K. Intestinal commensal microbes as immune modulators. Cell Host Microbe 2012, 12, 496–508.
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118.
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209.
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341.
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009, 31, 677–689.
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117.
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; van den Brandt, P.A.; Stobberingh, E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006, 118, 511–521.
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra265.
- Gritz, E.C.; Bhandari, V. The human neonatal gut microbiome: A brief review. Front. Pediatr. 2015, 3, 17.
- Sood, R.; Zehnder, J.L.; Druzin, M.L.; Brown, P.O. Gene expression patterns in human placenta. Proc. Natl. Acad. Sci. USA 2006, 103, 5478–5483.
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975.
- Pantoja-Feliciano, I.G.; Clemente, J.C.; Costello, E.K.; Perez, M.E.; Blaser, M.J.; Knight, R.; Dominguez-Bello, M.G. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 2013, 7, 1112–1115.
- Biasucci, G.; Rubini, M.; Riboni, S.; Morelli, L.; Bessi, E.; Retetangos, C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010, 86 (Suppl. S1), 13–15.
- Madan, J.C.; Farzan, S.F.; Hibberd, P.L.; Karagas, M.R. Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. Curr. Opin. Pediatr. 2012, 24, 753–759.
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313.
- Balmer, S.E.; Wharton, B.A. Diet and faecal flora in the newborn: Breast milk and infant formula. Arch. Dis. Child. 1989, 64, 1672–1677.
- Harmsen, H.J.; Wildeboer-Veloo, A.C.; Raangs, G.C.; Wagendorp, A.A.; Klijn, N.; Bindels, J.G.; Welling, G.W. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 61–67.
- Yoshioka, H.; Iseki, K.; Fujita, K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics 1983, 72, 317–321.
