The highly conserved and dynamically reversible N6-methyladenine (m6A) modification has emerged as a critical gene expression regulator by affecting RNA splicing, translation efficiency, and stability at the post-transcriptional level, which has been established to be involved in various physiological and pathological processes, including glycolipid metabolism and the development of glycolipid metabolic disease (GLMD). Hence, accumulating studies have focused on the effects and regulatory mechanisms of m6A modification on glucose metabolism, lipid metabolism, and GLMD. Glucose metabolism involves a very complex regulatory network, including anaerobic glycolysis, aerobic oxidation, pentose phosphate pathway, glycogen synthesis, and gluconeogenesis. An increasing number of studies have reported that m6A modification is an important regulatory mechanism of glucose homeostasis and downstream effects.
- N6-methyladenosine methylation
- glucose metabolism
- lipid metabolism
- glycolipid metabolic disease
1. Introduction
2. m6A Methylation
| Type | Regulator | Biological Function | References |
|---|---|---|---|
| m6A writers | METTL3 | Catalyzes m6A modification | [16] |
| METTL14 | Assists METTL3 to recognize the specific subtract and enhances the stability of MTC structure | [16] | |
| METTL16 | Catalyzes m6A modification and regulates SAM homeostasis | [17] | |
| WTAP | Promotes the cellular m6A deposition | [18] | |
| VIRMA (KIAA1429) |
Guides the core component of MTC to specific RNA region and interacts with cleavage and polyadenylation specific factor 5/6 (CPSF5/6) | [18] | |
| RBM15/15B | Recruits METTL3-METTL14 heterodimer to specific RNA sites | [19] | |
| ZC3H13 | Contributes to the nuclear localization of MTC | [20] | |
| IME4 | Mediates m6A modification of yeast mRNA | [21] | |
| MUM2 | Mediates m6A modification of yeast mRNA | [21] | |
| m6A erasers | FTO | Removes m6A modification to promote mRNA splicing and translation | [22] |
| ALKBH5 | Removes m6A modification to promote mRNA splicing, mRNA nuclear output, and long 3′-UTR mRNA production | [23] | |
| m6A readers | YTHDF1 | Promotes mRNA translation and protein synthesis | [24] |
| YTHDF2 | Reduces mRNA stability and regulates mRNA localization | [24] | |
| YTHDF3 | Interacts with YTHDF1 to promote mRNA translation or assists YTHDF2-mediated RNA degradation | [24] | |
| YTHDC1 | Contributes to RNA splicing, export, and transcriptional silencing | [25] | |
| YTHDC2 | Promotes the translation of target RNA but reduces their abundance | [26] | |
| eIF3 | Promotes mRNA translation | [27] | |
| IGF2BPs | Enhances the stability and translation of target RNA | [28] | |
| HNRNPA2B1 | Regulates alternative splicing and primary microRNA processing | [29] | |
| HNRNPC/G | Mediates mRNA splicing | [29] |
3. m6A Modification and Glucose Metabolism
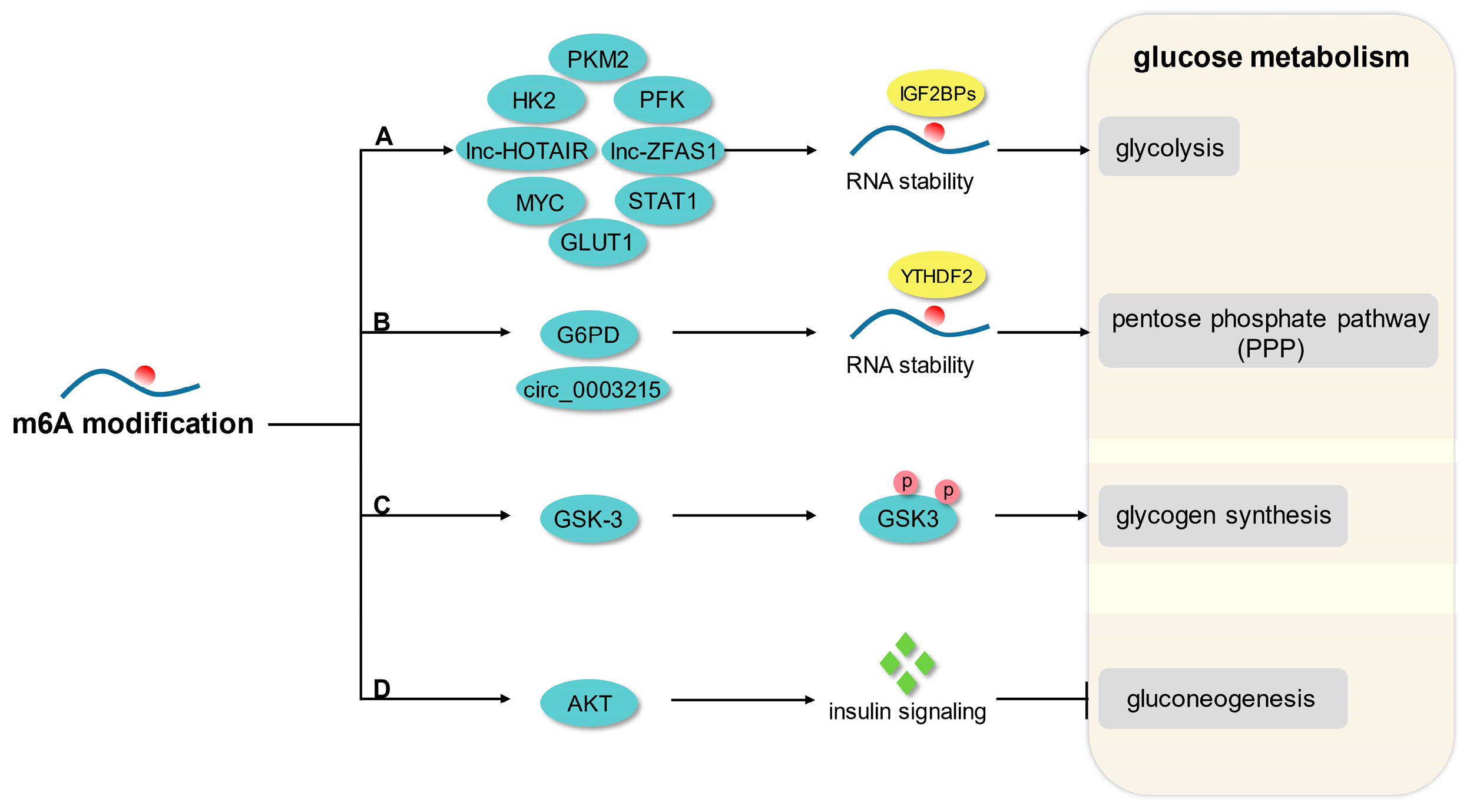
3.1. Glycolysis
3.2. Pentose Phosphate Pathway
3.3. Glycogen Synthesis and Gluconeogenesis
This entry is adapted from the peer-reviewed paper 10.3390/biom13020273
References
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887.
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29.
- Wei, C.; Gershowitz, A.; Moss, B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 1975, 4, 379–386.
- Bokar, J.; Shambaugh, M.; Polayes, D.; Matera, A.; Rottman, F. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247.
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176.
- Huang, H.; Weng, H.; Chen, J. m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 2020, 37, 270–288.
- Li, A.; Chen, Y.-S.; Ping, X.-L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.-Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447.
- Śledź, P.; Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. eLife 2016, 5, e18434.
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2013, 505, 117–120.
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519.
- Cai, Z.; Deng, X.; Zhao, L.; Wang, X.; Yang, L.; Yuan, G. The relationship between Schistosoma and glycolipid metabolism. Microb. Pathog. 2021, 159, 105120.
- Wu, J.; Frazier, K.; Zhang, J.; Gan, Z.; Wang, T.; Zhong, X. Emerging role of m6A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 2019, 21, e12942.
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206.
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646.
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641.
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317.
- Zhou, K.I.; Pan, T. Structures of the m6A Methyltransferase Complex: Two Subunits with Distinct but Coordinated Roles. Mol. Cell 2016, 63, 183–185.
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10.
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373.
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e6.
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-Resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis. Cell 2013, 155, 1409–1421.
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e5.
- Tang, C.; Klukovich, R.; Peng, H.; Wang, Z.; Yu, T.; Zhang, Y.; Zheng, H.; Klungland, A.; Yan, W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA 2017, 115, E325–E333.
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399.
- Roundtree, I.A.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 2017, 6, e31311.
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127.
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010.
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295, Correction in Nat. Cell Biol. 2018, 20, 1098; Correction in Nat. Cell Biol. 2020, 22, 1288.
- Wu, B.; Su, S.; Patil, D.P.; Liu, H.; Gan, J.; Jaffrey, S.R.; Ma, J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018, 9, 420.
- Boucheé, C.; Serdy, S.; Kahn, C.R.; Goldfine, A. The Cellular Fate of Glucose and Its Relevance in Type 2 Diabetes. Endocr. Rev. 2004, 25, 807–830.
- Butterfield, D.A.; Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019, 20, 148–160.
- Shen, C.; Xuan, B.; Yan, T.; Ma, Y.; Xu, P.; Tian, X.; Zhang, X.; Cao, Y.; Ma, D.; Zhu, X.; et al. m6A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer 2020, 19, 72.
- Zheng, Y.; Wang, Y.; Liu, Y.; Xie, L.; Ge, J.; Yu, G.; Zhao, G. N6-Methyladenosine Modification of PTTG3P Contributes to Colorectal Cancer Proliferation via YAP1. Front. Oncol. 2021, 11, 669731.
- Du, L.; Li, Y.; Kang, M.; Feng, M.; Ren, Y.; Dai, H.; Wang, Y.; Wang, Y.; Tang, B. USP48 Is Upregulated by Mettl14 to Attenuate Hepatocellular Carcinoma via Regulating SIRT6 Stabilization. Cancer Res. 2021, 81, 3822–3834.
- Lin, J.-X.; Lian, N.-Z.; Gao, Y.-X.; Zheng, Q.-L.; Yang, Y.-H.; Ma, Y.-B.; Xiu, Z.-S.; Qiu, Q.-Z.; Wang, H.-G.; Zheng, C.-H.; et al. m6A methylation mediates LHPP acetylation as a tumour aerobic glycolysis suppressor to improve the prognosis of gastric cancer. Cell Death Dis. 2022, 13, 463.
- Lyu, Y.; Zhang, Y.; Wang, Y.; Luo, Y.; Ding, H.; Li, P.; Ni, G. HIF-1α Regulated WTAP Overexpression Promoting the Warburg Effect of Ovarian Cancer by m6A-Dependent Manner. J. Immunol. Res. 2022, 2022, 6130806.
- Ou, B.; Liu, Y.; Yang, X.; Xu, X.; Yan, Y.; Zhang, J. C5aR1-positive neutrophils promote breast cancer glycolysis through WTAP-dependent m6A methylation of ENO1. Cell Death Dis. 2021, 12, 737.
- Yu, H.; Zhao, K.; Zeng, H.; Li, Z.; Chen, K.; Zhang, Z.; Li, E.; Wu, Z. N6-methyladenosine (m6A) methyltransferase WTAP accelerates the Warburg effect of gastric cancer through regulating HK2 stability. Biomed. Pharmacother. 2020, 133, 111075.
- Li, Y.; He, L.; Wang, Y.; Tan, Y.; Zhang, F. N6-methyladenosine methyltransferase KIAA1429 elevates colorectal cancer aerobic glycolysis via HK2-dependent manner. Bioengineered 2022, 13, 11923–11932.
- Yang, D.; Chang, S.; Li, F.; Ma, M.; Yang, J.; Lv, X.; Huangfu, L.; Jia, C. m6A transferase KIAA1429-stabilized LINC00958 accelerates gastric cancer aerobic glycolysis through targeting GLUT1. IUBMB Life 2021, 73, 1325–1333.
- Wang, Q.; Xie, H.; Peng, H.; Yan, J.; Han, L.; Ye, G. ZC3H13 Inhibits the Progression of Hepatocellular Carcinoma through m6A-PKM2-Mediated Glycolysis and Enhances Chemosensitivity. J. Oncol. 2021, 2021, 1328444.
- Sun, X.; Li, Q.; Yang, L. Sevoflurane Inhibits lncRNA HOTAIR-Modulated Stability of HK2 mRNA in a m6A-Dependent Manner to Dampen Aerobic Glycolysis and Proliferation in Lung Cancer. BioMed. Res. Int. 2022, 2022, 4668774.
- Wang, H.; Liang, Z.; Gou, Y.; Li, Z.; Cao, Y.; Jiao, N.; Tan, J.; Yu, Y.; Zhang, Z. FTO-dependent N(6)-Methyladenosine regulates the progression of endometriosis via the ATG5/PKM2 Axis. Cell. Signal. 2022, 98, 110406.
- Huang, J.; Sun, W.; Wang, Z.; Lv, C.; Zhang, T.; Zhang, D.; Dong, W.; Shao, L.; He, L.; Ji, X.; et al. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner. J. Exp. Clin. Cancer Res. 2022, 41, 42.
- Yang, X.; Shao, F.; Guo, D.; Wang, W.; Wang, J.; Zhu, R.; Gao, Y.; He, J.; Lu, Z. WNT/β-catenin-suppressed FTO expression increases m6A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis. Cell Death Dis. 2021, 12, 462.
- Qing, Y.; Dong, L.; Gao, L.; Li, C.; Li, Y.; Han, L.; Prince, E.; Tan, B.; Deng, X.; Wetzel, C.; et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol. Cell 2021, 81, 922–939.e9.
- Yu, H.; Yang, X.; Tang, J.; Si, S.; Zhou, Z.; Lu, J.; Han, J.; Yuan, B.; Wu, Q.; Lu, Q.; et al. ALKBH5 Inhibited Cell Proliferation and Sensitized Bladder Cancer Cells to Cisplatin by m6A-CK2α-Mediated Glycolysis. Mol. Ther. Nucleic. Acids 2020, 23, 27–41.
- Lu, S.; Han, L.; Hu, X.; Sun, T.; Xu, D.; Li, Y.; Chen, Q.; Yao, W.; He, M.; Wang, Z.; et al. N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: Implication in colorectal cancer. J. Hematol. Oncol. 2021, 14, 188.
- Hu, C.; Liu, T.; Han, C.; Xuan, Y.; Jiang, D.; Sun, Y.; Zhang, X.; Zhang, W.; Xu, Y.; Liu, Y.; et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m6A-MYC expression. Int. J. Biol. Sci. 2022, 18, 507–521.
- Luo, F.; Lin, K. N6-methyladenosine (m6A) reader IGF2BP1 accelerates gastric cancer aerobic glycolysis in c-Myc-dependent manner. Exp. Cell Res. 2022, 417, 113176.
- Wang, Y.; Lu, J.-H.; Wu, Q.-N.; Jin, Y.; Wang, D.-S.; Chen, Y.-X.; Liu, J.; Luo, X.-J.; Meng, Q.; Pu, H.-Y.; et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer 2019, 18, 174.
- Li, Z.; Peng, Y.; Li, J.; Chen, Z.; Chen, F.; Tu, J.; Lin, S.; Wang, H. N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat. Commun. 2020, 11, 2578.
- Huangfu, N.; Zheng, W.; Xu, Z.; Wang, S.; Wang, Y.; Cheng, J.; Li, Z.; Cheng, K.; Zhang, S.; Chen, X.; et al. RBM4 regulates M1 macrophages polarization through targeting STAT1-mediated glycolysis. Int. Immunopharmacol. 2020, 83, 106432.
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.-M.; Krüger, A.; Tauqeer Alam, M.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963.
- Clasquin, M.F.; Melamud, E.; Singer, A.; Gooding, J.R.; Xu, X.; Dong, A.; Cui, H.; Campagna, S.R.; Savchenko, A.; Yakunin, A.F.; et al. Riboneogenesis in Yeast. Cell 2011, 145, 969–980.
- Wang, L.; Xie, J.; Schultz, P.G. Expanding the Genetic Code. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 225–249.
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197.
- Liu, Z.; Chen, Y.; Wang, L.; Ji, S. ALKBH5 Promotes the Proliferation of Glioma Cells via Enhancing the mRNA Stability of G6PD. Neurochem. Res. 2021, 46, 3003–3011.
- Sheng, H.; Li, Z.; Su, S.; Sun, W.; Zhang, X.; Li, L.; Li, J.; Liu, S.; Lu, B.; Zhang, S.; et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinog. 2019, 41, 541–550.
- Chen, B.; Hong, Y.; Gui, R.; Zheng, H.; Tian, S.; Zhai, X.; Xie, X.; Chen, Q.; Qian, Q.; Ren, X.; et al. N6-methyladenosine modification of circ_0003215 suppresses the pentose phosphate pathway and malignancy of colorectal cancer through the miR-663b/DLG4/G6PD axis. Cell Death Dis. 2022, 13, 804.
- Han, H.-S.; Kang, G.; Kim, J.S.; Choi, B.H.; Koo, S.-H. Regulation of glucose metabolism from a liver-centric perspective. Exp. Mol. Med. 2016, 48, e218.
- Li, H.; Zhang, N.; Jiao, X.; Wang, C.; Sun, W.; He, Y.; Ren, G.; Huang, S.; Li, M.; Chang, Y.; et al. Downregulation of microRNA-6125 promotes colorectal cancer growth through YTHDF2-dependent recognition of N6-methyladenosine-modified GSK3β. Clin. Transl. Med. 2021, 11, e602.
- Zhou, L.; Jiang, J.; Huang, Z.; Jin, P.; Peng, L.; Luo, M.; Zhang, Z.; Chen, Y.; Na Xie, N.; Gao, W.; et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m6A-mediated degradation of STEAP3 mRNA. Mol. Cancer 2022, 21, 168.
- Zou, Y.; Zheng, S.; Xie, X.; Ye, F.; Hu, X.; Tian, Z.; Yan, S.-M.; Yang, L.; Kong, Y.; Tang, Y.; et al. N6-methyladenosine regulated FGFR4 attenuates ferroptotic cell death in recalcitrant HER2-positive breast cancer. Nat. Commun. 2022, 13, 2672.
- Diao, M.-Y.; Zhu, Y.; Yang, J.; Xi, S.-S.; Wen, X.; Gu, Q.; Hu, W. Hypothermia protects neurons against ischemia/reperfusion-induced pyroptosis via m6A-mediated activation of PTEN and the PI3K/Akt/GSK-3β signaling pathway. Brain Res. Bull. 2020, 159, 25–31.
- Gao, X.-Q.; Zhang, Y.-H.; Liu, F.; Ponnusamy, M.; Zhao, X.-M.; Zhou, L.-Y.; Zhai, M.; Liu, C.-Y.; Li, X.-M.; Wang, M.; et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N6-methyladenosine methylation of Parp10 mRNA. Nature 2020, 22, 1319–1331.
- Hatting, M.; Tavares, C.D.; Sharabi, K.; Rines, A.K.; Puigserver, P. Insulin regulation of gluconeogenesis. Ann. N. Y. Acad. Sci. 2017, 1411, 21–35.
- Liu, J.; Luo, G.; Sun, J.; Men, L.; Ye, H.; He, C.; Ren, D. METTL14 is essential for β-cell survival and insulin secretion. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2019, 1865, 2138–2148.
