CTC is the main target of liquid biopsy. In the past few decades, the separation of CTC based on the electrochemical method has attracted widespread attention due to its convenience, rapidness, low cost, high sensitivity, and no need for complex instruments and equipment. In this review, we summarized the latest developments in the electrochemical-based CTC detection, and discussed the challenges and possible trends.
- circulating tumor cells
- electrochemical detection
- direct assays
- sandwich assays
1. Introduction
Cancer is one of the leading causes of death, and around 90% cancer death due to metastasis [1][2]. Therefore, achieving an earlier cancer diagnosis is of fundamental importance. For conventional needle biopsy techniques, the invasiveness limits its use. Meanwhile, liquid biopsy techniques analyze tumor cells or tumor cell debris from blood or other body fluids, including circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), extracellular vesicles (EVs), and exosomes, etc. [3]. Compared with needle biopsy, its non-invasiveness allows us to collect patient blood samples continuously, and to realize real-time monitoring of patient disease progression and personalized medicine [4][5]. Moreover, since CTC, ctDNA, EVs, etc. can be released from both primary and metastatic tumors, liquid biopsy provides us more comprehensive information [6][7]. CTC is the main target of liquid biopsy, for CTC is the most important part during the metastasis process [8]. It has been reported that CTC could be detected before cancer forms metastasis [9][10]. Detection of CTC in the blood could be used to achieve an earlier diagnosis and a better control of cancer, and avoid the bad consequences caused by cancer metastasis. Besides, CTC could be used to assess the patient prognosis and evaluate the treatment outcome in real-time [5][11]. The isolation, culture, and sequencing of CTC could also help us to determine patients’ drug resistance and find potential therapeutic targets [12][13][14][15].
Separating CTC from complex blood components is extremely challenging. The amount of CTC in the blood is extremely rare, in average about 1–100 mL-1 [16], while the number of white blood cells (WBC) and red blood cells (RBC) is about 0.4–1×107 mL-1 and 3.5–5×109 mL-1, respectively. The separation mainly relies on the difference in biological properties or physical properties between CTC and blood cells [17]. Biological properties-based CTC separation, mainly using the unique antigen expression on the surface of CTC, such as the most commonly used anti-epithelial cell adhesion molecule (EpCAM) and Cytokeratin (CK), etc. [18]. Coupling these antibodies to the surface of magnetic beads or the chip can achieve the specific capture and separation of CTC. Physical properties-based CTC separation mainly uses the difference in cell density, size, and deformability between CTC and blood cells to achieve CTC separation [19][20][21]. Although there are many CTC detection methods, their complicated operation process, high cost, and low sensitivity are still problems. In recent years, a lot of electrochemical methods have also been used to detect CTC, using aptamers and nanomaterials to modify the electrode, by recording the current change/electrical impedance spectrum change, and establishing a linear relationship between the change and the number of CTC to realize the quantification of CTC [22][23]. This ensures high sensitivity and selectivity, and has outstanding advantages, such as rapid response, easy operation, affordability, and nondestructive analysis [22].
After completing the separation of CTC, it is very important to quantify the number of CTC. In general, traditional biological properties-based methods and physical properties-based methods use fluorescently labeled antibodies to identify and count the captured CTCs. CTCs were recognized as Hoechst+ (nuclear dye), EpCAM+/CK+, and CD45- (WBC specific marker) cells, while WBCs were recognized as Hoechst+, EpCAM-/CK-, and CD45+ cells [24][25][26]. This fluorescence imaging-based method usually requires a specialized fluorescence microscope, which is expensive, and the output of the results requires professional technicians and also takes a long time, thus limiting its clinical use. The electrochemical-based method only requires some simple instruments, such as current meters, and the whole detection process is relatively simple and fast [27][28][29][30]. However, the preparation process of the device is complicated and the detection time is long. There is an urgent need for a simpler, more efficient, and faster method. Point-of-care testing (POCT) realizes target quantification through pressure, distance, color, etc. As a sensitive, fast, cheap, easy-to-operate method, it allows patients to realize sample input and result output, and it doesn’t need for complex equipment [31][32]. The test can be performed at the bedside of the patient and the results are available immediately. It is of great significance for popularizing the clinical application of CTC detection.
2. Electrochemical Detection of CTC
In recent years, electrochemical detection of CTC has attracted widespread attention due to its advantages such as convenience, rapidity, low cost, high sensitivity, low detection limit, and strong specificity [33]. From the principle of separation, electrochemical detection of CTC is a kind of the biological properties-based CTC separation. However, it usually uses aptamer to replace traditional antibodies. Aptamer has similar specificity and affinity compared to antibodies, but it has many advantages, including easier to achieve large-scale production, lower cost, and lower immunogenicity [22][34] This greatly reduces the cost of electrochemical detection.
The principle of electrochemical detection is to modify the electrode using nanomaterials and cancer cell specific aptamer, and CTC is bonded to aptamer (on the electrode surface), which will cause the current/electrochemiluminescence (ECL)/photoelectrochemical (PEC) signal to decrease or EC impedance spectroscopy (EIS) signal to increase [27][35][36][37][38][39]. By establishing a linear relationship between the change and the number of CTC, the quantification can be realized. The electrochemical CTC detection methods can be roughly divided into direct assays, sandwich assays and other assays [22]. The direct assays use nanomaterials-aptamer as the capture probe (which is directly modified on the electrode) to bind and detect CTC [40]. The sandwich assays add a signal probe on the basis of direct assays, that is, after CTC is bound to the capture probe on the electrode, a signal probe (another kind of nanomaterials-aptamer) is added. After the signal probe is combined with CTC, it can significantly amplify the signal, thereby increasing the sensitivity and reducing the detection limit [30][41][42]. We summarized the electrochemical-based CTC detection methods in Table 1, including the direct assay and the sandwich assay, and we listed the assay time, linear range, limit of detection, capture probe, and signal probe.
2.1. Direct Assays
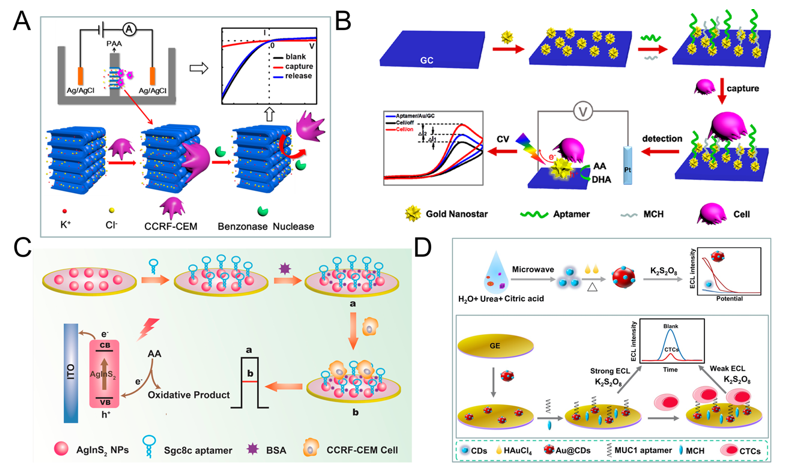
The principle of direct assays is based on the combination of CTC and electrode, thus affects the transfer of electrons, which in turn leads to a decrease in current/increase in electrical impedance [37][43], thereby realizing the quantitative detection of CTC. Many researchers use functionalized nanomaterials to modify the electrodes, the main purpose is to increase the contact area, enhance the conductivity, and then enhance the signal. Cao et al. proposed a method for detecting acute leukemia CCRF-CEM cells using a hybrid of nanochannels and ion channels arrays. The main principle is after modifying the sgc8c aptamer on the surface of the channel, CTC is combined with ion channel, then it will dramatically block the ionic flow through channels, which in turn causes current changes, using linear sweep voltammetry (LSV) cytosensor to record the change and to quantify CTC. Compared with a single channel, the array channels could amplify the signal and to improve the detection sensitivity. The linear range is 1×102–2×106 cells mL-1, and the limit of detection (LOD) is 100 cells mL-1 (Figure 1A) [44]. Subsequently, Li et al. reported a PEC-based CTC detection method, hypotoxic ternary mercaptopropionic acid (MPA)-capped AgInS2 nanoparticles (NPs) as PEC sensing substrates, after being excited by red light, it shows high photon-current conversion efficiency, and generates strong photocurrent. After CCRF-CEM cell is connected to aptamers on AgInS2 NPs, the photocurrent is significantly reduced, and CTC can be detected. Its linear range is from 1.5×102 to 3.0×105 cells mL-1, and the LOD is 16 cells mL-1 (Figure 1C) [36]. In order to further improve the detection sensitivity, Wang et al. prepared nanomaterials-plasmonic gold nanostars (AuNSs), modified sgc8c aptamer on its surface and fixed it on glassy carbon (GC) electrode. The results showed that the LOD for CCRF-CEM cell is as low as 5 cells mL-1, in addition, MCF-7 cells could also be detected with a LOD of 10 cells mL-1 (Figure 1B) [45]. Liu et al. used ECL cytosensor for MCF-7 cell detection. Modified Au@CDs-aptamer on GC electrode, after adding K2S2O8, Au@CDs showed strong electrochemiluminescence. Due to the combination of CTC, the strength of ECL will decrease. Its linear range was 100 to 10,000 cells mL-1 with a LOD of 34 cells mL-1 (Figure 1D) [35]. Graphene nanomaterials are also widely used in electrochemical detection of CTC due to their excellent electrical conductivity [46]. For example, Zhang et al. synthetized a xFe2O3-nPt (or nPt-xFe2O3)-coated graphene nanostructure (xFe2O3-nPt@graphene) to detect MCF-7 cells using differential pulse voltammetry (DPV) cytosensor. The combination of graphene, Pt and Fe2O3 greatly reduced the electrode resistance and improved the electron transfer efficiency, and effectively amplified the signal and improved the sensitivity. The linear range is 18 to 1.5 × 106 cells mL-1, and the LOD is 6 cells mL-1 [47]. Bábelová et al. invented a method to detect leukemia, which is based on electrochemical impedance spectroscopy (EIS) and thickness shear mode acoustic method (TSM). It used sgc8c as the specific aptamer and achieved high-sensitivity detection of Jurkat cell. The LOD of electrochemical and acoustic sensors is 105±10 and 463±50 cells mL-1, respectively [48].
Table 1. Summary of electrochemical CTC detection methods.
|
Assay Time (min) |
Linear Range (cells mL-1) |
LOD (cells mL-1) |
Aptamer/Antibody |
Target Cell |
Capture Probe |
Signal Probe |
Clinical Sample |
Ref. |
|
|
Direct assay |
70 |
18-1.5×106 |
6 |
AS1411 |
MCF-7 |
xFe2O3-nPt (or nPt-xFe2O3)-coated graphene nanostructure+aptamer |
—— |
Spiked blood samples |
[47] |
|
45 |
1×102-2×106 |
100 |
sgc8c |
CCRF-CEM |
Nanochannel−ion channel +aptamer |
—— |
—— |
[44] |
|
|
90 |
100-10,000 |
34 |
anti-MUC 1 aptamer |
MCF-7 |
Gold electrode+Au@CDs +aptamer |
—— |
Human serum samples |
[35] |
|
|
45 |
5-1×105 |
5 |
sgc8c |
CCRF-CEM |
Glassy carbon electrode +AuNSs+aptamer |
—— |
Human serum samples and spiked blood samples |
[45] |
|
|
60 |
1.5×102-3×105 |
16 |
sgc8c |
CCRF-CEM |
AgInS2 NPs-modified electrode+aptamer |
—— |
—— |
[36] |
|
|
Sandwich assay |
120 |
75-5,500 |
75 |
E-cadherin |
Not limited |
CNT-AuNPs+antibody |
E-cadherin antibody+QD |
Fetal calf serum and mouse blood |
[49] |
|
50 |
1×102-5×104 |
5 |
SYL3C |
MCF-7 |
m-aptamer/MCH/AuE |
Anti-EpCAM/HRP-AuNP |
Spiked blood samples |
[28] |
|
|
20 |
5-500 |
4 |
Td05 |
Ramos |
AuNPs-Fe3O4-GS+aptamer |
AuNPs with the electroactive species (Fc-SH/Thi) +aptamer |
Spiked blood samples and 3 leukemia patients |
[41] |
|
|
3 |
Sgc8 |
CCRF-CEM |
|||||||
|
40 |
10-1×106 |
2 |
EpCAM+vimentin |
MCF-7 |
Ketjen black+AuNP +antibody |
PdIrBPMNS+antibody |
Spiked blood samples |
[29] |
|
|
170 |
3-3,000 |
1 |
5’-thiol modified MCF-7 binding aptamer |
MCF-7 |
MN+EpCAM |
Cu2O+aptamer |
Spiked blood samples |
[30] |
|
|
60 |
3-1,000 |
1 |
anti-MUC 1 aptamer |
MCF-7 |
MN+EpCAM |
LiFePO4/Au+aptamer |
Spiked blood samples |
[27] |
|
|
115 |
5-3×104 |
1 |
SYL3C-RCA primer |
MCF-7 |
MN+EpCAM |
Aptamer+DNA amplification (RCA) |
Spiked blood samples |
[42] |
|
|
40 |
20-1×106 |
3 |
EpCAM |
MCF-7 |
Au electrode+AB +AuNP+proteinG+antibody |
Pt@AgNFs+antibody |
Spiked blood samples |
[37] |
Figure 1. Direct electrochemical detection of CTC. (A) Linear sweep voltammetry (LSV) cytosensor for CCRF-CEM cell detection, using hybrid nanochannels and ion channels. Reprinted with permission from ref [44]. Copyright 2017, American Chemical Society. (B) Cyclic voltammetry (CV) cytosensor for CCRF-CEM cell and MCF-7 cell detection, by modifying plasmonic gold nanostars (AuNSs)-aptamer on glassy carbon (GC) electrodes. Reprinted with permission from ref [45]. Copyright 2019, American Chemical Society. (C) Photoelectrochemical (PEC) cytosensor for CCRF-CEM cell detection, using AgInS2 nanoparticles (NPs) coupled with sgc8c aptamer which exhibited high photon-to-current conversion efficiency under red light excitation. Reprinted with permission from ref [36]. Copyright 2019, Elsevier. (D) Electrochemiluminescence (ECL) cytosensor for MCF-7 cell detection using Au@CDs-aptamer, after adding K2S2O8, Au@CDs showed strong electrochemiluminescence. Reprinted with permission from ref [35]. Copyright 2020, Elsevier.
2.2. Sandwich Assays
The sandwich assays add a signal probe on the basis of direct assays. The capture probe is fixed on the electrodes to identify and capture CTC. The signal probe is used for amplifying the signal, improving the sensitivity, and reducing the detection limit [27][37][50]. Signal probes are usually some active nanomaterials or particles with enzymatic activity.
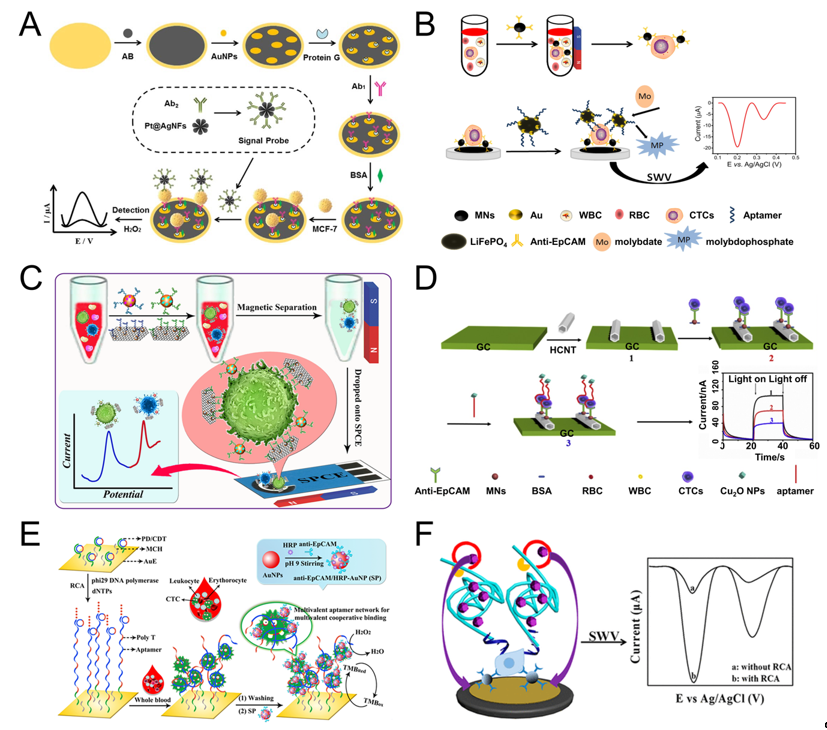
Shen et al. used magnetic nanospheres modified with anti-EpCAM antibody to capture MCF-7 cells. The captured MCF-7 cells are spread on the electrode surface, aptamer (SYL3C-RCA primer) is added and bound to the cells, by rolling circle amplification (RCA), producing a large amount of DNA molecules and reacting with the substrate to amplify the electrochemical current signal. The linear range is 5-3×104 mL-1, and the detection limit can reach 1 cell mL-1 (Figure 2F) [42]. RCA technology was also used by other groups [28][51]. For example, Yang et al. used RCA reaction to expand the initial PD/CDT so that the expanded DNA chain contains multiple duplicate aptamers. The expanded DNA strand as a capture probe has the following advantages: (1) This flexible long DNA chain can extend to the depth of the cell suspension, which allows the aptamers in full contact with the target cells; (2) The aptamer binds to the target cell in a multivalent manner, effectively improve the capture efficiency of target cells. The signal probe anti-EpCAM/Horseradish peroxidase (HRP)-gold nanoparticles (AuNP) is used to catalyze H2O2 to generate electrical signals for detection. The linear range is 1 × 102-5×104 mL-1, and the LOD is 5 cell mL-1 (Figure 2E) [28].
Tang et al. modified AuNPs/Acetylene black (AuNPs/AB) on the gold electrode to increase the specific surface area and enhance the conductivity of the gold electrode. Then anchored antibody on the surface of AuNPs/AB to capture MCF-7 cells. After capture, the signal probe Pt@AgNFs was added, which can catalyze H2O2 to generate electrical signals. The linear range is 20-1×106, and the detection limit is 3 cells mL-1 (Figure 2A) [37]. Luo et al. proposed a PEC biosensor-based CTC detection method with higher sensitivity using antibody-magnetic nanospheres to capture MCF-7 cells. After spreading the capture cells on the electrode, the photocurrent intensity of hexagonal carbon nitride tubes (HCNT) is reduced due to the steric hindrance derived from MCF-7. After adding a signal probe Cu2O NPs, the photocurrent intensity of was further decreased because Cu2O NPs competitively absorbed the excitation light, and the aptamer molecules further increased the steric hindrance, the linear range is 3–3000 cell mL-1, and the detection limit is 1 cell mL-1 (Figure 2D) [30]. Similarly, Zhang et al. also used antibody-magnetic nanospheres to capture MCF-7 cells, and after coating the cells on the electrode, used gold nanoparticle-modified LiFePO4 coupled with the aptamer as a signal probe, due to the reaction of phosphate group in LiFePO4 with molybdate that formed redox molybdophosphate (PMo12O40) precipitates and caused current change. The linear range is 3–1000 cells mL-1, and the LOD is 1 cell mL-1 (Figure 2B) [27]. The above parts are all about the detection of one type of cell. Using multiple different aptamers can simultaneously detect multiple types of cells. For example, Dou et al. used AuNPs-Fe3O4-graphene nanosheets (GS) as the capture probe, AuNPs coupled with aptamer (Td05/Sgc8) and decorated with the electroactive species (Fc-SH/Thi) as the signal probe, and simultaneously achieved the quantitative detection of Ramos and CCRF-CEM cells, the detection limit is 4 and 3 cells mL-1, respectively. Further, the linear range is 5–500 cells mL-1. More importantly, they also performed testing of three leukemia patients and showed obvious results (Figure 2C) [41]. Du et al. proposed a Cyclic voltammetry (CV) cytosensor based CTC electrochemical detection, and it can specifically detect the change of E-cadherin and analyze different stages of EMT. They modified CNT-AuNPs on the electrode surface, and then used quantum dot (QD) coupled anti E-cadherin antibody to bind to CTC. QD itself acts as a signal probe to enhance electrical signal, and at the same time, QD can also provide a strong fluorescent signal for imaging [49].
In Table 1, we can see that, compared to the direct method, sandwich assays have higher sensitivity, and its LOD can be as low as 1 cell mL-1. However, the detection time may be longer. In addition, the fabrication and detection process are also more complicated. The common problems are the narrow range of applicable cell types and very few clinical trials.
Figure 2. Sandwich electrochemical detection of CTC. (A) Voltammetric cytosensor for differential pulse voltammetry (DPV) detection of MCF-7 cells, using AuNPs/Acetylene black-antibody as capture probe and Pt@Ag nanoflowers-antibody as signal probe. Reprinted with permission from ref [37]. Copyright 2018, Elsevier. (B) Voltammetric cytosensor for square wave voltammetry (SWV) detection of MCF-7 cells, anti-EpCAM antibody-modified Fe3O4 magnetic nanospheres (MNs) were used to capture CTC, and gold nanoparticles modified LiFePO4 (LiFePO4/Au) particles were used as signal probe. Reprinted with permission from ref [27]. Copyright 2020, Elsevier. (C) Voltammetric cytosensor for SWV detection of CCRF-CEM cells, Au nanoparticles (AuNP) array-decorated magnetic graphene nanosheet as capture probe and aptamer/electroactive species-loaded AuNP as signal probe. Reprinted with permission from ref [41]. Copyright 2019, American Chemical Society. (D) Photoelectrochemical (PEC) cytosensor for detection of MCF-7 cells, Magnetic Fe3O4 nanospheres (MNs) as capture probe and Cu2O nanoparticles as signal probe. Reprinted with permission from ref [30], Copyright 2020, Elsevier. (E) Cyclic voltammetry (CV) cytosensor for MCF-7 cell detection, via rolling circle amplification (RCA) extension of the electrode-immobilized primer/circular DNA complexes as the capture probe, and anti-EpCAM/HRP-AuNP as signal probe. Reprinted with permission from ref [28]. Copyright 2020, American Chemical Society. (F) Voltammetric cytosensor for SWV detection of MCF-7 cells, anti-EpCAM antibody-modified magnetic nanospheres were used to capture and enrich CTC, and aptamer−primer DNA sequence as a signal probe, by RCA, produced a large amount of DNA molecules and reacted with the substrate to amplify the signal. Reprinted with permission from ref [42]. Copyright 2019, American Chemical Society.
This entry is adapted from the peer-reviewed paper 10.3390/s20216073
References
- Steeg, P.S. Targeting metastasis. Rev. Cancer 2016, 16, 201–218, doi:10.1038/nrc.2016.25
- Sleeman, J.; Steeg, P.S. Cancer metastasis as a therapeutic target. J. Cancer 2010, 46, 1177–1180, doi:10.1016/j.ejca.2010.02.039.
- Vaidyanathan, R.; Soon, R.H.; Zhang, P.; Jiang, K.; Lim, C.T. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab Chip.2018, 19, 11–34, doi:10.1039/c8lc00684a.
- Tadimety, A.; Closson, A.; Li, C.; Yi, S.; Shen, T.; Zhang, J.X.J. Advances in liquid biopsy on-chip for cancer management: Technologies, biomarkers, and clinical analysis. Rev. Clin. Lab. Sci. 2018, 55, 140–162, doi:10.1080/10408363.2018.1425976.
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Cancer Res. 2006, 12, 4218–4224, doi:10.1158/1078-0432.CCR-05-2821.
- Snow, A.; Chen, D.; Lang, J.E. The current status of the clinical utility of liquid biopsies in cancer. Expert Rev. Mol. Diagn. 2019, 19, 1031–1041, doi:10.1080/14737159.2019.1664290.
- Tellez-Gabriel, M.; Heymann, M.F.; Heymann, D. Circulating Tumor Cells as a Tool for Assessing Tumor Heterogeneity. Theranostics 2019, 9, 4580–4594, doi:10.7150/thno.34337.
- O’Flaherty, J.D.; Gray, S.; Richard, D.; Fennell, D.; O’Leary, J.J.; Blackhall, F.H.; O’Byrne, K.J. Circulating tumour cells, their role in metastasis and their clinical utility in lung cancer. Lung Cancer 2012, 76, 19–25, doi:10.1016/j.lungcan.2011.10.018.
- Samlowski, W.E.; McGregor, J.R.; Samlowski, S.T.; Tharkar, S.; Shen, S.; Bentz, J.S. Growth of Circulating Tumor Cell-Derived Colonies from Peripheral Blood of Melanoma Patients: Preliminary Characterization of Colony Composition. Health 2014, 6, 1467–1481, doi:10.4236/health.2014.612181.
- Cheung, K.J.; Ewald, A.J. A collective route to metastasis: Seeding by tumor cell clusters. Science 2016, 352, 167–169, doi:10.1126/science.aaf6546.
- Wang, C.; Mu, Z.; Chervoneva, I.; Austin, L.; Ye, Z.; Rossi, G.; Palazzo, J.P.; Sun, C.; Abu-Khalaf, M.; Myers, R.E.; et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res. Treat. 2017, 161, 83–94, doi:10.1007/s10549-016-4026-2.
- Khoo, B.L.; Grenci, G.; Lim, Y.B.; Lee, S.C.; Han, J.; Lim, C.T. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Protoc. 2018, 13, 34–58, doi:10.1038/nprot.2017.125.
- Yin, S.; Xi, R.; Wu, A.; Wang, S.; Li, Y.; Wang, C.; Tang, L.; Xia, Y.; Yang, D.; Li, J.; et al. Patient-derived tumor-like cell clusters for drug testing in cancer therapy. Transl. Med. 2020, 12, doi:10.1126/scitranslmed.aaz1723.
- Zhou, J.; Tu, C.; Liang, Y.; Huang, B.; Fang, Y.; Liang, X.; Ye, X. The label-free separation and culture of tumor cells in a microfluidic biochip. Analyst 2020, 145, 1706–1715, doi:10.1039/c9an02092f.
- Cheng, Y.H.; Chen, Y.C.; Lin, E.; Brien, R.; Jung, S.; Chen, Y.T.; Lee, W.; Hao, Z.; Sahoo, S.; Min Kang, H.; et al. Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Commun. 2019, 10, 2163, doi:10.1038/s41467-019-10122-2.
- Hou, H.W.; Warkiani, M.E.; Khoo, B.L.; Li, Z.R.; Soo, R.A.; Tan, D.S.; Lim, W.T.; Han, J.; Bhagat, A.A.; Lim, C.T. Isolation and retrieval of circulating tumor cells using centrifugal forces. Rep. 2013, 3, 1259, doi:10.1038/srep01259.
- Alix-Panabieres, C.; Pantel, K. Challenges in circulating tumour cell research. Rev. Cancer 2014, 14, 623–631, doi:10.1038/nrc3820.
- Weissenstein, U.; Schumann, A.; Reif, M.; Link, S.; Toffol-Schmidt, U.D.; Heusser, P. Detection of circulating tumor cells in blood of metastatic breast cancer patients using a combination of cytokeratin and EpCAM antibodies. BMC Cancer 2012, 12, 206, doi:10.1186/1471-2407-12-206.
- Banko, P.; Lee, S.Y.; Nagygyorgy, V.; Zrinyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. Hematol. Oncol. 2019, 12, 48, doi:10.1186/s13045-019-0735-4.
- Lin, Z.; Luo, G.; Du, W.; Kong, T.; Liu, C.; Liu, Z. Recent Advances in Microfluidic Platforms Applied in Cancer Metastasis: Circulating Tumor Cells’ (CTCs) Isolation and Tumor-On-A-Chip. Small 2020, 16, e1903899, doi:10.1002/smll.201903899.
- Nasiri, R.; Shamloo, A.; Ahadian, S.; Amirifar, L.; Akbari, J.; Goudie, M.J.; Lee, K.; Ashammakhi, N.; Dokmeci, M.R.; Di Carlo, D.; et al. Microfluidic-Based Approaches in Targeted Cell/Particle Separation Based on Physical Properties: Fundamentals and Applications. Small 2020, 16, e2000171, doi:10.1002/smll.202000171.
- Sun, D.; Lu, J.; Zhang, L.; Chen, Z. Aptamer-based electrochemical cytosensors for tumor cell detection in cancer diagnosis: A review. Chim. Acta 2019, 1082, 1–17, doi:10.1016/j.aca.2019.07.054.
- Tang, Z.; Huang, J.; He, H.; Ma, C.; Wang, K. Contributing to liquid biopsy: Optical and electrochemical methods in cancer biomarker analysis. Chem. Rev. 2020, 415, doi:10.1016/j.ccr.2020.213317.
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239, doi:10.1038/nature06385.
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Natl. Acad. Sci. USA 2010, 107, 18392–18397, doi:10.1073/pnas.1012539107.
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Methods 2015, 12, 685–691, doi:10.1038/nmeth.3404.
- Zhang, W.; Chen, H.; Yang, M.; Liao, L. Electrochemical assay for detection of circulating tumor cells based on LiFePO4 as electrochemical probe. Lett. 2020, 276, doi:10.1016/j.matlet.2020.128219.
- Yang, J.; Li, X.; Jiang, B.; Yuan, R.; Xiang, Y. In Situ-Generated Multivalent Aptamer Network for Efficient Capture and Sensitive Electrochemical Detection of Circulating Tumor Cells in Whole Blood. Chem. 2020, 92, 7893–7899, doi:10.1021/acs.analchem.0c01195.
- Peng, Y.; Peng, Y.; Tang, S.; Shen, H.; Sheng, S.; Wang, Y.; Wang, T.; Cai, J.; Xie, G.; Feng, W. PdIrBP mesoporous nanospheres combined with superconductive carbon black for the electrochemical determination and collection of circulating tumor cells. Acta 2020, 187, 216, doi:10.1007/s00604-020-4213-z.
- Luo, J.; Liang, D.; Zhao, D.; Yang, M. Photoelectrochemical detection of circulating tumor cells based on aptamer conjugated Cu2O as signal probe. Bioelectron. 2020, 151, 111976, doi:10.1016/j.bios.2019.111976.
- Park, J.; Han, D.H.; Park, J.K. Towards practical sample preparation in point-of-care testing: User-friendly microfluidic devices. Lab Chip 2020, 20, 1191–1203, doi:10.1039/d0lc00047g.
- Liu, D.; Tian, T.; Chen, X.; Lei, Z.; Song, Y.; Shi, Y.; Ji, T.; Zhu, Z.; Yang, L.; Yang, C. Gas-generating reactions for point-of-care testing. Analyst 2018, 143, 1294–1304, doi:10.1039/c8an00011e.
- Zhang, Z.; Li, Q.; Du, X.; Liu, M. Application of electrochemical biosensors in tumor cell detection. Thorac. Cancer 2020, 11, 840–850.
- Safarpour, H.; Dehghani, S.; Nosrati, R.; Zebardast, N.; Alibolandi, M.; Mokhtarzadeh, A.; Ramezani, M. Optical and electrochemical-based nano-aptasensing approaches for the detection of circulating tumor cells (CTCs). Biosens. Bioelectron. 2020, 148, 111833.
- Liu, P.; Wang, L.; Zhao, K.; Liu, Z.; Cao, H.; Ye, S.; Liang, G. High luminous efficiency Au@CDs for sensitive and label-free electrochemiluminescent detection of circulating tumor cells in serum. Sens. Actuators B Chem. 2020, 316.
- Li, J.; Lin, X.; Zhang, Z.; Tu, W.; Dai, Z. Red light-driven photoelectrochemical biosensing for ultrasensitive and scatheless assay of tumor cells based on hypotoxic AgInS2 nanoparticles. Biosens. Bioelectron. 2019, 126, 332–338.
- Tang, S.; Shen, H.; Hao, Y.; Huang, Z.; Tao, Y.; Peng, Y.; Guo, Y.; Xie, G.; Feng, W. A novel cytosensor based on Pt@Ag nanoflowers and AuNPs/Acetylene black for ultrasensitive and highly specific detection of Circulating Tumor Cells. Biosens. Bioelectron. 2018, 104, 72–78.
- Peng, Y.; Pan, Y.; Han, Y.; Sun, Z.; Jalalah, M.; Al-Assiri, M.S.; Harraz, F.A.; Yang, J.; Li, G. Direct Analysis of Rare Circulating Tumor Cells in Whole Blood Based on Their Controlled Capture and Release on Electrode Surface. Anal. Chem. 2020.
- Cao, H.-X.; Liu, P.-F.; Wang, L.; Liu, Z.-J.; Ye, S.-Y.; Liang, G.-X. Nonenzymatic chemiluminescence detection of circulating tumor cells in blood based on Au@luminol nanoparticles, hybridization chain reaction and magnetic isolation. Sens. Actuators B Chem. 2020, 318.
- Vajhadin, F.; Ahadian, S.; Travas-Sejdic, J.; Lee, J.; Mazloum-Ardakani, M.; Salvador, J.; Aninwene, G.E., 2nd; Bandaru, P.; Sun, W.; Khademhossieni, A. Electrochemical cytosensors for detection of breast cancer cells. Biosens. Bioelectron. 2020, 151, 111984.
- Dou, B.; Xu, L.; Jiang, B.; Yuan, R.; Xiang, Y. Aptamer-Functionalized and Gold Nanoparticle Array-Decorated Magnetic Graphene Nanosheets Enable Multiplexed and Sensitive Electrochemical Detection of Rare Circulating Tumor Cells in Whole Blood. Anal. Chem. 2019, 91, 10792–10799.
- Shen, C.; Liu, S.; Li, X.; Yang, M. Electrochemical Detection of Circulating Tumor Cells Based on DNA Generated Electrochemical Current and Rolling Circle Amplification. Anal. Chem. 2019, 91, 11614–11619.
- Shen, H.; Yang, J.; Chen, Z.; Chen, X.; Wang, L.; Hu, J.; Ji, F.; Xie, G.; Feng, W. A novel label-free and reusable electrochemical cytosensor for highly sensitive detection and specific collection of CTCs. Biosens. Bioelectron. 2016, 81, 495–502.
- Cao, J.; Zhao, X.P.; Younis, M.R.; Li, Z.Q.; Xia, X.H.; Wang, C. Ultrasensitive Capture, Detection, and Release of Circulating Tumor Cells Using a Nanochannel-Ion Channel Hybrid Coupled with Electrochemical Detection Technique. Anal. Chem. 2017, 89, 10957–10964.
- Wang, S.S.; Zhao, X.P.; Liu, F.F.; Younis, M.R.; Xia, X.H.; Wang, C. Direct Plasmon-Enhanced Electrochemistry for Enabling Ultrasensitive and Label-Free Detection of Circulating Tumor Cells in Blood. Anal. Chem. 2019, 91, 4413–4420.
- Cordaro, A.; Neri, G.; Sciortino, M.T.; Scala, A.; Piperno, A. Graphene-Based Strategies in Liquid Biopsy and in Viral Diseases Diagnosis. Nanomaterials 2020, 10, 1014.
- Zhang, H.; Liang, F.; Wu, X.; Liu, Y.; Chen, A. Recognition and sensitive detection of CTCs using a controllable label-free electrochemical cytosensor. Mikrochim. Acta 2020, 187, 487.
- Bábelová, L.; Sohová, M.E.; Poturnayová, A.; Buríková, M.; Bizík, J.; Hianik, T. Label-free Electrochemical Aptasensor for Jurkat Cells Detection as a Potential Diagnostic Tool for Leukemia. Electroanalysis 2018, 30, 1487–1495.
- Du, X.; Zhang, Z.; Zheng, X.; Zhang, H.; Dong, D.; Zhang, Z.; Liu, M.; Zhou, J. An electrochemical biosensor for the detection of epithelial-mesenchymal transition. Nat. Commun. 2020, 11, 192.
- Zhou, X.; Li, Y.; Wu, H.; Huang, W.; Ju, H.; Ding, S. A amperometric immunosensor for sensitive detection of circulating tumor cells using a tyramide signal amplification-based signal enhancement system. Biosens. Bioelectron. 2019, 130, 88–94.
- Sun, D.; Lu, J.; Luo, Z.; Zhang, L.; Liu, P.; Chen, Z. Competitive electrochemical platform for ultrasensitive cytosensing of liver cancer cells by using nanotetrahedra structure with rolling circle amplification. Biosens. Bioelectron. 2018, 120, 8–14.