Oncogenic mutations within the epidermal growth factor receptor (EGFR) kinase domain are well-established driver mutations in non–small cell lung cancer (NSCLC). Small-molecule tyrosine kinase inhibitors (TKIs) specifically targeting these mutations have improved treatment outcomes for patients with this subtype of NSCLC. Multiple targeted agents, including first-, second- and third-generation EGFR TKIs, have been approved or are under active investigation for patients with EGFR-mutant NSCLC. The first-generation TKIs, including erlotinib and gefitinib, are reversible inhibitors, binding both to mutant as well as wild-type (WT) EGFR. The second-generation TKIs, including afatinib and dacomitinib, are irreversible inhibitors that covalently bind to EGFR. The third-generation EGFR TKIs, including osimertinib (AZD9291), aumolertinib (HS-10296) and alflutinib (AST2818), are irreversible inhibitors that selectively bind to mutant EGFR and show greater efficacy than the first- and second-generation TKIs. Osimertinib has been approved by the FDA for both frontline and second-line treatment of NSCLC with EGFR-sensitizing mutations. Aumolertinib (HS-10296) and alflutinib (AST2818) have been approved for the treatment of EGFR-mutant NSCLC in China. Acquired resistance inevitably occurs, and a promising new generation of EGFR-targeting agents is under investigation.
- non–small cell lung cancer
- EGFR
- structure
- targeted therapy
1. Classical Mutations
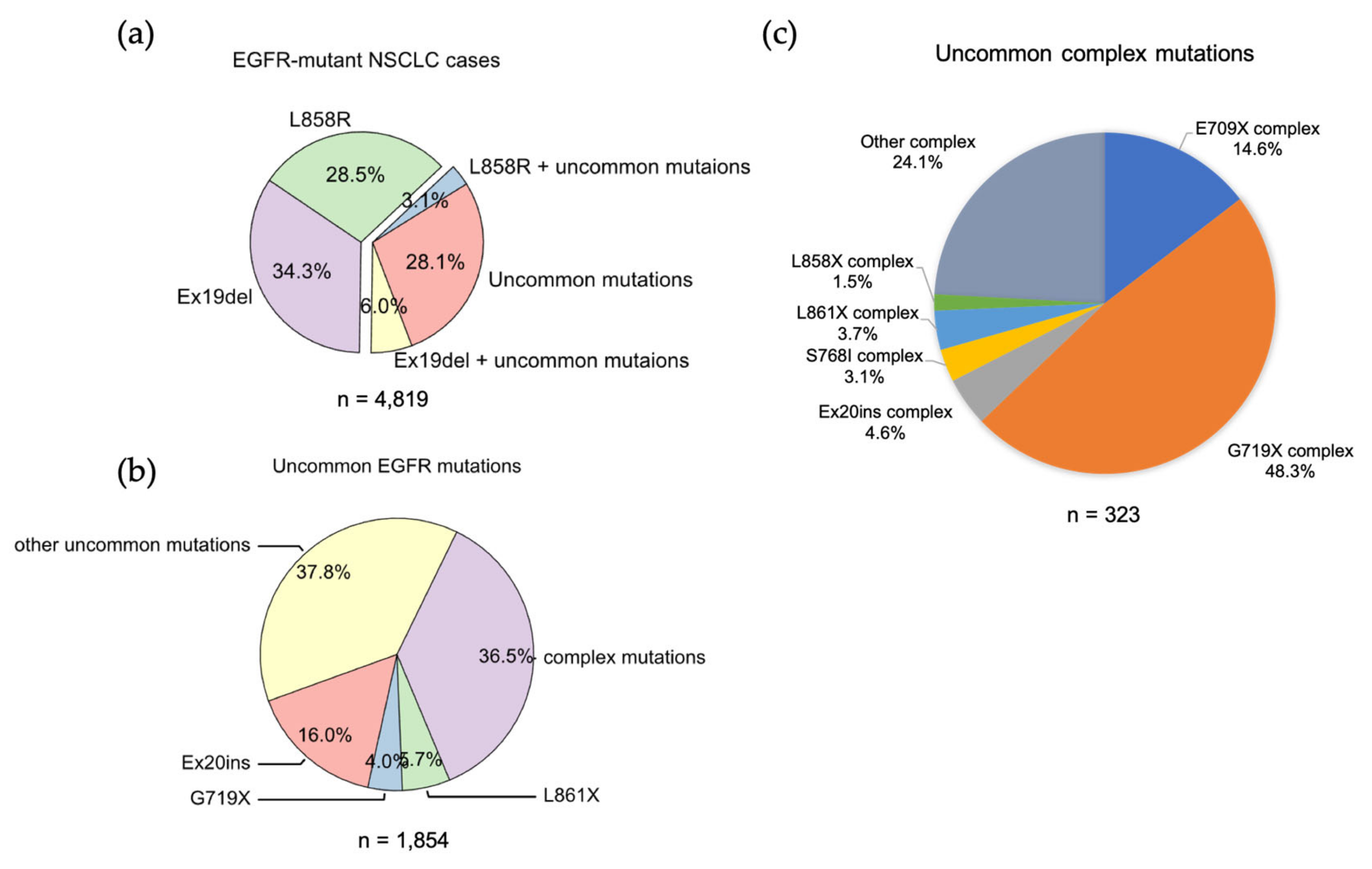
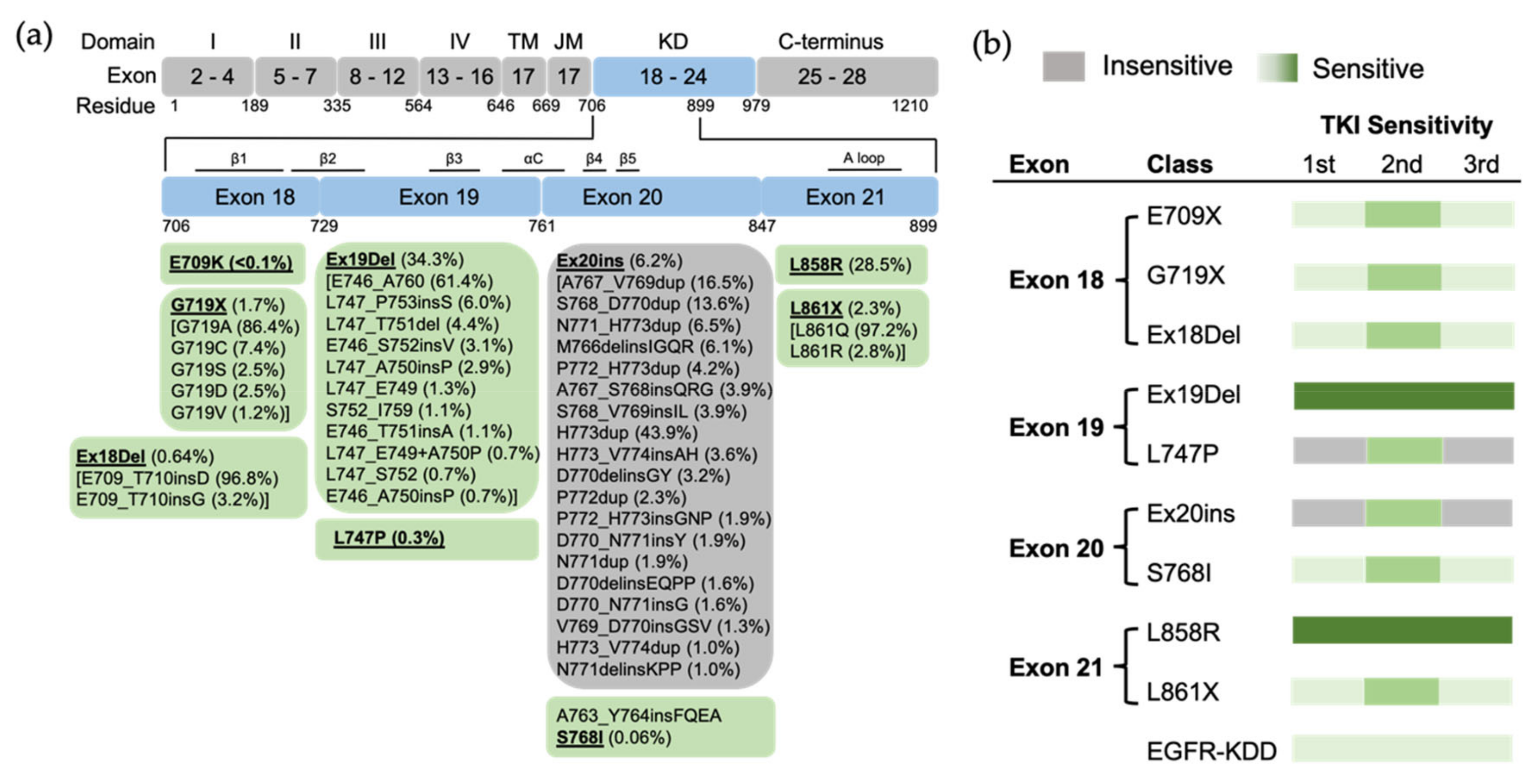
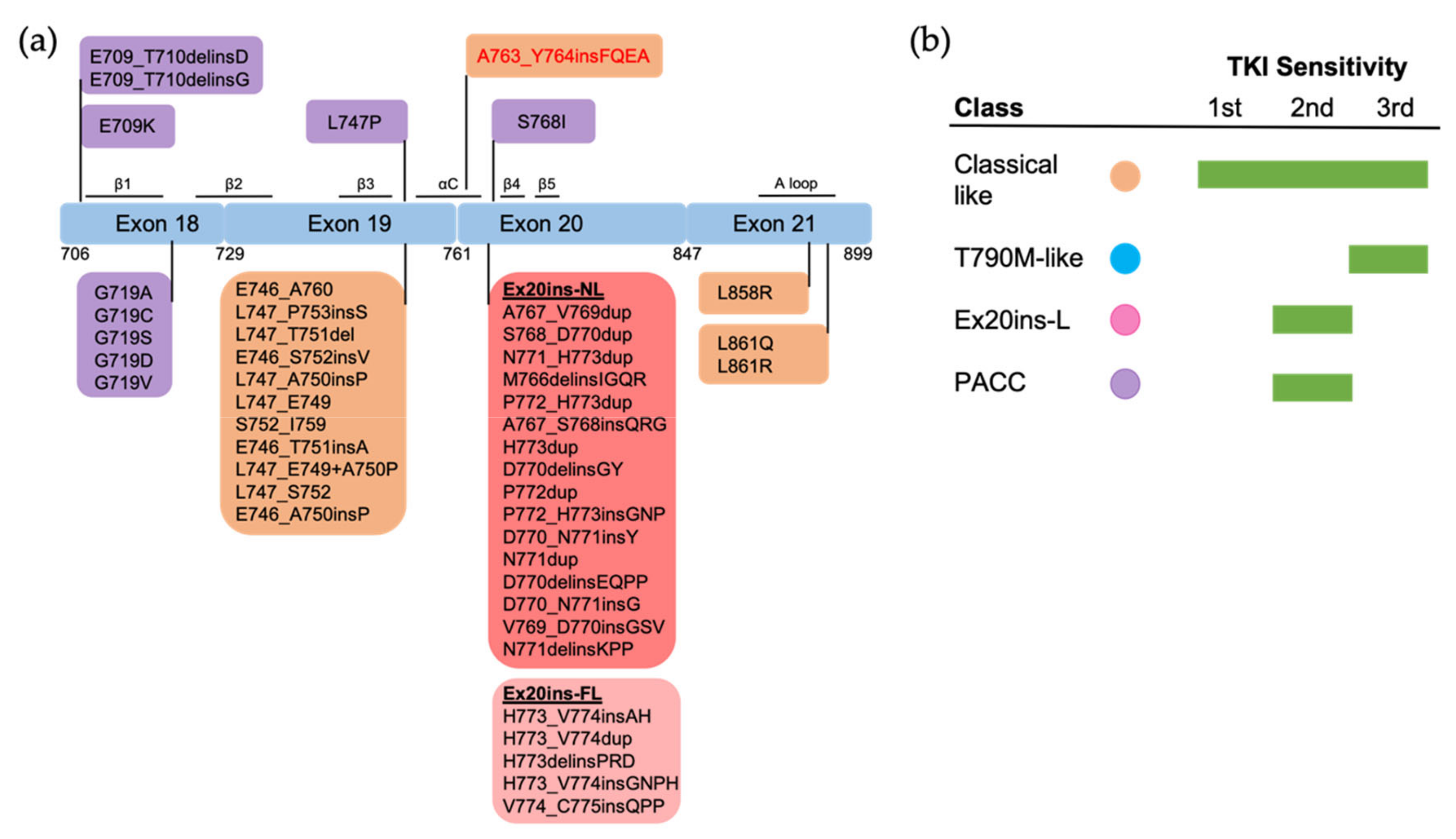
2. Uncommon EGFR Mutations
2.1. Exon 18 Mutations
2.2. Exon 19 Mutations
2.3. Exon 20 Mutations
2.4. Exon 21 Mutations
2.5. EGFR Kinase Domain Duplication
2.6. Uncommon Complex Mutations
| E709X | Partner Mutation | Count | G719X | Partner Mutation | Count |
|---|---|---|---|---|---|
| E709A | G719A | 12 | G719C | S768I | 43 |
| E709K | G719A | 11 | G719A | S768I | 24 |
| E709A | G719R | 8 | G719S | S768I | 14 |
| E709A | G719C | 3 | G719A | L861Q | 12 |
| E709K | G719S | 3 | G719S | L861Q | 11 |
| E709K | G719C | 2 | G719A | R776H | 6 |
| E709V | G719C | 2 | G719A | L833V | 5 |
| E709V | G719S | 1 | G719S | S768N | 3 |
| E709A | L861R | 1 | G719S | R776H | 3 |
| E709A | L861Q | 1 | G719D | L861Q | 2 |
| Ex20ins | Partner Mutation | Count | L861X | Partner Mutation | Count |
|---|---|---|---|---|---|
| N771_H773dup | V845L | 2 | L861Q | S720F | 3 |
| D770delinsEQPL | H773Y | 1 | L861Q | S768I | 2 |
| D770delinsEL | N771Y | 1 | L861R | S768I | 1 |
| D770_V774dup | Q791E | 1 | L861R | P1073T | 1 |
| D770_P772dup | H773Y | 1 | L861Q | R776H | 1 |
| D770_N771insY | S1030L | 1 | L861Q | R776C | 1 |
| D770_N771insG | N771Y | 1 | L861Q | L838V | 1 |
| A767_V769dup | R836C | 1 | L861Q | L833F | 1 |
| A767_S768insQRG | V765L | 1 | L861Q | L747F | 1 |
| H773delinsQI | N771H | 1 |
This entry is adapted from the peer-reviewed paper 10.3390/biom13020210
References
- Mok, T.S.; Wu, Y.L.; Thongprasert, S.; Yang, C.H.; Chu, D.T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957.
- Yun, C.H.; Boggon, T.J.; Li, Y.; Woo, M.S.; Greulich, H.; Meyerson, M.; Eck, M.J. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 2007, 11, 217–227.
- Foster, S.A.; Whalen, D.M.; Ozen, A.; Wongchenko, M.J.; Yin, J.; Yen, I.; Schaefer, G.; Mayfield, J.D.; Chmielecki, J.; Stephens, P.J.; et al. Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2. Cancer Cell 2016, 29, 477–493.
- Lu, S.; Wang, Q.; Zhang, G.; Dong, X.; Yang, C.T.; Song, Y.; Chang, G.C.; Lu, Y.; Pan, H.; Chiu, C.H.; et al. Efficacy of Aumolertinib (HS-10296) in Patients with Advanced EGFR T790M+ NSCLC: Updated Post-National Medical Products Administration Approval Results from the APOLLO Registrational Trial. J. Thorac. Oncol. 2022, 17, 411–422.
- Shi, Y.; Zhang, S.; Hu, X.; Feng, J.; Ma, Z.; Zhou, J.; Yang, N.; Wu, L.; Liao, W.; Zhong, D.; et al. Safety, Clinical Activity, and Pharmacokinetics of Alflutinib (AST2818) in Patients with Advanced NSCLC With EGFR T790M Mutation. J. Thorac. Oncol. 2020, 15, 1015–1026.
- Zhou, W.; Ercan, D.; Chen, L.; Yun, C.H.; Li, D.; Capelletti, M.; Cortot, A.B.; Chirieac, L.; Iacob, R.E.; Padera, R.; et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 2009, 462, 1070–1074.
- Schuler, M.; Yang, J.C.; Park, K.; Kim, J.H.; Bennouna, J.; Chen, Y.M.; Chouaid, C.; De Marinis, F.; Feng, J.F.; Grossi, F.; et al. Afatinib beyond progression in patients with non-small-cell lung cancer following chemotherapy, erlotinib/gefitinib and afatinib: Phase III randomized LUX-Lung 5 trial. Ann. Oncol. 2016, 27, 417–423.
- Bazhenova, L.; Minchom, A.; Viteri, S.; Bauml, J.M.; Ou, S.I.; Gadgeel, S.M.; Trigo, J.M.; Backenroth, D.; Li, T.; Londhe, A.; et al. Comparative clinical outcomes for patients with advanced NSCLC harboring EGFR exon 20 insertion mutations and common EGFR mutations. Lung Cancer 2021, 162, 154–161.
- He, M.; Capelletti, M.; Nafa, K.; Yun, C.H.; Arcila, M.E.; Miller, V.A.; Ginsberg, M.S.; Zhao, B.; Kris, M.G.; Eck, M.J.; et al. EGFR exon 19 insertions: A new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin. Cancer Res. 2012, 18, 1790–1797.
- Baek, J.H.; Sun, J.M.; Min, Y.J.; Cho, E.K.; Cho, B.C.; Kim, J.H.; Ahn, M.J.; Park, K. Efficacy of EGFR tyrosine kinase inhibitors in patients with EGFR-mutated non-small cell lung cancer except both exon 19 deletion and exon 21 L858R: A retrospective analysis in Korea. Lung Cancer 2015, 87, 148–154.
- Kohsaka, S.; Nagano, M.; Ueno, T.; Suehara, Y.; Hayashi, T.; Shimada, N.; Takahashi, K.; Suzuki, K.; Takamochi, K.; Takahashi, F.; et al. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci. Transl. Med. 2017, 9, eaan6566.
- Xu, J.; Jin, B.; Chu, T.; Dong, X.; Yang, H.; Zhang, Y.; Wu, D.; Lou, Y.; Zhang, X.; Wang, H.; et al. EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non-small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: A real-world study in China. Lung Cancer 2016, 96, 87–92.
- Shan, Y.; Eastwood, M.P.; Zhang, X.; Kim, E.T.; Arkhipov, A.; Dror, R.O.; Jumper, J.; Kuriyan, J.; Shaw, D.E. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell 2012, 149, 860–870.
- Red Brewer, M.; Yun, C.H.; Lai, D.; Lemmon, M.A.; Eck, M.J.; Pao, W. Mechanism for activation of mutated epidermal growth factor receptors in lung cancer. Proc. Natl. Acad. Sci. USA 2013, 110, E3595–E3604.
- Robichaux, J.P.; Le, X.; Vijayan, R.S.K.; Hicks, J.K.; Heeke, S.; Elamin, Y.Y.; Lin, H.Y.; Udagawa, H.; Skoulidis, F.; Tran, H.; et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2021, 597, 732–737.
- Tamirat, M.Z.; Koivu, M.; Elenius, K.; Johnson, M.S. Structural characterization of EGFR exon 19 deletion mutation using molecular dynamics simulation. PLoS ONE 2019, 14, e0222814.
- Brown, B.P.; Zhang, Y.K.; Kim, S.; Finneran, P.; Yan, Y.; Du, Z.; Kim, J.; Hartzler, A.L.; LeNoue-Newton, M.L.; Smith, A.W.; et al. Allele-specific activation, enzyme kinetics, and inhibitor sensitivities of EGFR exon 19 deletion mutations in lung cancer. Proc. Natl. Acad. Sci. USA 2022, 119, e2206588119.
- Truini, A.; Starrett, J.H.; Stewart, T.; Ashtekar, K.; Walther, Z.; Wurtz, A.; Lu, D.; Park, J.H.; DeVeaux, M.; Song, X.; et al. The EGFR Exon 19 Mutant L747-A750>P Exhibits Distinct Sensitivity to Tyrosine Kinase Inhibitors in Lung Adenocarcinoma. Clin. Cancer Res. 2019, 25, 6382–6391.
- Zhao, C.; Jiang, T.; Li, J.; Wang, Y.; Su, C.; Chen, X.; Ren, S.; Li, X.; Zhou, C. The impact of EGFR exon 19 deletion subtypes on clinical outcomes in non-small cell lung cancer. Transl. Lung Cancer Res. 2020, 9, 1149–1158.
- Yun, C.H.; Mengwasser, K.E.; Toms, A.V.; Woo, M.S.; Greulich, H.; Wong, K.K.; Meyerson, M.; Eck, M.J. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA 2008, 105, 2070–2075.
- Yu, H.A.; Pao, W. Targeted therapies: Afatinib--new therapy option for EGFR-mutant lung cancer. Nat. Rev. Clin. Oncol. 2013, 10, 551–552.
- Yan, X.E.; Ayaz, P.; Zhu, S.J.; Zhao, P.; Liang, L.; Zhang, C.H.; Wu, Y.C.; Li, J.L.; Choi, H.G.; Huang, X.; et al. Structural Basis of AZD9291 Selectivity for EGFR T790M. J. Med. Chem. 2020, 63, 8502–8511.
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125.
- Cho, B.C.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Sriuranpong, V.; Imamura, F.; Nogami, N.; Kurata, T.; Okamoto, I.; Zhou, C.; et al. Osimertinib versus Standard of Care EGFR TKI as First-Line Treatment in Patients with EGFRm Advanced NSCLC: FLAURA Asian Subset. J. Thorac. Oncol. 2019, 14, 99–106.
- Yasuda, H.; Ichihara, E.; Sakakibara-Konishi, J.; Zenke, Y.; Takeuchi, S.; Morise, M.; Hotta, K.; Sato, M.; Matsumoto, S.; Tanimoto, A.; et al. A phase I/II study of osimertinib in EGFR exon 20 insertion mutation-positive non-small cell lung cancer. Lung Cancer 2021, 162, 140–146.
- Kobayashi, Y.; Mitsudomi, T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci. 2016, 107, 1179–1186.
- Bar, J.; Peled, N.; Schokrpur, S.; Wolner, M.; Rotem, O.; Girard, N.; Aboubakar Nana, F.; Derijcke, S.; Kian, W.; Patel, S.; et al. UNcommon EGFR Mutations: International Case Series on Efficacy of Osimertinib in Real-Life Practice in First-LiNe Setting (UNICORN). J. Thorac. Oncol. 2022; in press.
- Kobayashi, Y.; Togashi, Y.; Yatabe, Y.; Mizuuchi, H.; Jangchul, P.; Kondo, C.; Shimoji, M.; Sato, K.; Suda, K.; Tomizawa, K.; et al. EGFR Exon 18 Mutations in Lung Cancer: Molecular Predictors of Augmented Sensitivity to Afatinib or Neratinib as Compared with First- or Third-Generation TKIs. Clin. Cancer Res. 2015, 21, 5305–5313.
- Minari, R.; Leonetti, A.; Gnetti, L.; Zielli, T.; Ventura, L.; Bottarelli, L.; Lagrasta, C.; La Monica, S.; Petronini, P.G.; Alfieri, R.; et al. Afatinib therapy in case of EGFR G724S emergence as resistance mechanism to osimertinib. Anticancer Drugs 2021, 32, 758–762.
- Li, Y.; Lin, Y.; Wu, J.; Ye, F. Meningeal metastasis patients with EGFR G724S who develop resistance to osimertinib benefit from the addition of afatinib. Transl. Lung Cancer Res. 2020, 9, 2188–2190.
- Fang, W.; Huang, Y.; Gan, J.; Zheng, Q.; Zhang, L. Emergence of EGFR G724S After Progression on Osimertinib Responded to Afatinib Monotherapy. J. Thorac. Oncol. 2020, 15, e36–e37.
- Wei, Y.; Jiang, B.; Liu, S.; Zhang, Z.; Fang, W.; Yang, Y.; Li, X.; Zhao, J.; Zhao, H. Afatinib as a Potential Therapeutic Option for Patients with NSCLC With EGFR G724S. JTO Clin. Res. Rep. 2021, 2, 100193.
- Wang, Y.T.; Ning, W.W.; Li, J.; Huang, J.A. Exon 19 L747P mutation presented as a primary resistance to EGFR-TKI: A case report. J. Thorac. Dis. 2016, 8, E542–E546.
- Huang, J.; Wang, Y.; Zhai, Y.; Wang, J. Non-small cell lung cancer harboring a rare EGFR L747P mutation showing intrinsic resistance to both gefitinib and osimertinib (AZD9291): A case report. Thorac. Cancer 2018, 9, 745–749.
- Li, J.; Zhu, L.; Stebbing, J.; Peng, L. Afatinib treatment in a lung adenocarcinoma patient harboring a rare EGFR L747P mutation. J. Cancer Res. Ther. 2022, 18, 1436–1439.
- Li, Y.; Guo, W.; Jiang, B.; Han, C.; Ye, F.; Wu, J. Case Report: Dacomitinib is effective in lung adenocarcinoma with rare EGFR mutation L747P and brain metastases. Front. Oncol. 2022, 12, 863771.
- Yoshizawa, T.; Uchibori, K.; Araki, M.; Matsumoto, S.; Ma, B.; Kanada, R.; Seto, Y.; Oh-Hara, T.; Koike, S.; Ariyasu, R.; et al. Microsecond-timescale MD simulation of EGFR minor mutation predicts the structural flexibility of EGFR kinase core that reflects EGFR inhibitor sensitivity. NPJ Precis. Oncol. 2021, 5, 32.
- Meador, C.B.; Sequist, L.V.; Piotrowska, Z. Targeting EGFR Exon 20 Insertions in Non-Small Cell Lung Cancer: Recent Advances and Clinical Updates. Cancer Discov. 2021, 11, 2145–2157.
- Friedlaender, A.; Subbiah, V.; Russo, A.; Banna, G.L.; Malapelle, U.; Rolfo, C.; Addeo, A. EGFR and HER2 exon 20 insertions in solid tumours: From biology to treatment. Nat. Rev. Clin. Oncol. 2022, 19, 51–69.
- Lin, Y.T.; Shih, J.Y. Not All EGFR Exon 20 Insertions Are Created Equal. JTO Clin. Res. Rep. 2020, 1, 100069.
- Shi, C.; Xing, R.; Li, M.; Feng, J.; Sun, R.; Wei, B.; Guo, Y.; Ma, J.; Wang, H. Real-world clinical treatment outcomes in Chinese non-small cell lung cancer with EGFR exon 20 insertion mutations. Front. Oncol. 2022, 12, 949304.
- Yang, J.C.; Sequist, L.V.; Geater, S.L.; Tsai, C.M.; Mok, T.S.; Schuler, M.; Yamamoto, N.; Yu, C.J.; Ou, S.H.; Zhou, C.; et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838.
- Beau-Faller, M.; Prim, N.; Ruppert, A.M.; Nanni-Metellus, I.; Lacave, R.; Lacroix, L.; Escande, F.; Lizard, S.; Pretet, J.L.; Rouquette, I.; et al. Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10 117 patients: A multicentre observational study by the French ERMETIC-IFCT network. Ann. Oncol. 2014, 25, 126–131.
- Naidoo, J.; Sima, C.S.; Rodriguez, K.; Busby, N.; Nafa, K.; Ladanyi, M.; Riely, G.J.; Kris, M.G.; Arcila, M.E.; Yu, H.A. Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: Clinical outcomes and response to erlotinib. Cancer 2015, 121, 3212–3220.
- Janning, M.; Suptitz, J.; Albers-Leischner, C.; Delpy, P.; Tufman, A.; Velthaus-Rusik, J.L.; Reck, M.; Jung, A.; Kauffmann-Guerrero, D.; Bonzheim, I.; et al. Treatment outcome of atypical EGFR mutations in the German National Network Genomic Medicine Lung Cancer (nNGM). Ann. Oncol. 2022, 33, 602–615.
- Yasuda, H.; Park, E.; Yun, C.H.; Sng, N.J.; Lucena-Araujo, A.R.; Yeo, W.L.; Huberman, M.S.; Cohen, D.W.; Nakayama, S.; Ishioka, K.; et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci. Transl. Med. 2013, 5, 216ra177.
- Vasconcelos, P.; Gergis, C.; Viray, H.; Varkaris, A.; Fujii, M.; Rangachari, D.; VanderLaan, P.A.; Kobayashi, I.S.; Kobayashi, S.S.; Costa, D.B. EGFR-A763_Y764insFQEA Is a Unique Exon 20 Insertion Mutation That Displays Sensitivity to Approved and In-Development Lung Cancer EGFR Tyrosine Kinase Inhibitors. JTO Clin. Res. Rep. 2020, 1, 100051.
- Voon, P.J.; Tsui, D.W.; Rosenfeld, N.; Chin, T.M. EGFR exon 20 insertion A763-Y764insFQEA and response to erlotinib--Letter. Mol. Cancer Ther. 2013, 12, 2614–2615.
- Qin, Y.; Jian, H.; Tong, X.; Wu, X.; Wang, F.; Shao, Y.W.; Zhao, X. Variability of EGFR exon 20 insertions in 24 468 Chinese lung cancer patients and their divergent responses to EGFR inhibitors. Mol. Oncol. 2020, 14, 1695–1704.
- Kobayashi, I.S.; Viray, H.; Rangachari, D.; Kobayashi, S.S.; Costa, D.B. EGFR-D770>GY and Other Rare EGFR Exon 20 Insertion Mutations with a G770 Equivalence Are Sensitive to Dacomitinib or Afatinib and Responsive to EGFR Exon 20 Insertion Mutant-Active Inhibitors in Preclinical Models and Clinical Scenarios. Cells 2021, 10, 3561.
- Ruan, Z.; Kannan, N. Altered conformational landscape and dimerization dependency underpins the activation of EGFR by alphaC-beta4 loop insertion mutations. Proc. Natl. Acad. Sci. USA 2018, 115, E8162–E8171.
- Elamin, Y.Y.; Robichaux, J.P.; Carter, B.W.; Altan, M.; Tran, H.; Gibbons, D.L.; Heeke, S.; Fossella, F.V.; Lam, V.K.; Le, X.; et al. Poziotinib for EGFR exon 20-mutant NSCLC: Clinical efficacy, resistance mechanisms, and impact of insertion location on drug sensitivity. Cancer Cell 2022, 40, 754–767.e6.
- Udagawa, H.; Hasako, S.; Ohashi, A.; Fujioka, R.; Hakozaki, Y.; Shibuya, M.; Abe, N.; Komori, T.; Haruma, T.; Terasaka, M.; et al. TAS6417/CLN-081 Is a Pan-Mutation-Selective EGFR Tyrosine Kinase Inhibitor with a Broad Spectrum of Preclinical Activity against Clinically Relevant EGFR Mutations. Mol. Cancer Res. 2019, 17, 2233–2243.
- Gonzalvez, F.; Vincent, S.; Baker, T.E.; Gould, A.E.; Li, S.; Wardwell, S.D.; Nadworny, S.; Ning, Y.; Zhang, S.; Huang, W.S.; et al. Mobocertinib (TAK-788): A Targeted Inhibitor of EGFR Exon 20 Insertion Mutants in Non-Small Cell Lung Cancer. Cancer Discov. 2021, 11, 1672–1687.
- Leal, J.L.; Alexander, M.; Itchins, M.; Wright, G.M.; Kao, S.; Hughes, B.G.M.; Pavlakis, N.; Clarke, S.; Gill, A.J.; Ainsworth, H.; et al. EGFR Exon 20 Insertion Mutations: Clinicopathological Characteristics and Treatment Outcomes in Advanced Non-Small Cell Lung Cancer. Clin. Lung Cancer 2021, 22, e859–e869.
- Popat, S.; Hsia, T.C.; Hung, J.Y.; Jung, H.A.; Shih, J.Y.; Park, C.K.; Lee, S.H.; Okamoto, T.; Ahn, H.K.; Lee, Y.C.; et al. Tyrosine Kinase Inhibitor Activity in Patients with NSCLC Harboring Uncommon EGFR Mutations: A Retrospective International Cohort Study (UpSwinG). Oncologist 2022, 27, 255–265.
- Hellmann, M.D.; Reva, B.; Yu, H.; Rusch, V.W.; Rizvi, N.A.; Kris, M.G.; Arcila, M.E. Clinical and in vivo evidence that EGFR S768I mutant lung adenocarcinomas are sensitive to erlotinib. J. Thorac. Oncol. 2014, 9, e73–e74.
- Leventakos, K.; Kipp, B.R.; Rumilla, K.M.; Winters, J.L.; Yi, E.S.; Mansfield, A.S. S768I Mutation in EGFR in Patients with Lung Cancer. J. Thorac. Oncol. 2016, 11, 1798–1801.
- Kuiper, J.L.; Hashemi, S.M.; Thunnissen, E.; Snijders, P.J.; Grunberg, K.; Bloemena, E.; Sie, D.; Postmus, P.E.; Heideman, D.A.; Smit, E.F. Non-classic EGFR mutations in a cohort of Dutch EGFR-mutated NSCLC patients and outcomes following EGFR-TKI treatment. Br. J. Cancer 2016, 115, 1504–1512.
- Niogret, J.; Coudert, B.; Boidot, R. Primary Resistance to Afatinib in a Patient with Lung Adenocarcinoma Harboring Uncommon EGFR Mutations: S768I and V769L. J. Thorac. Oncol. 2018, 13, e113.
- Cho, J.H.; Lim, S.H.; An, H.J.; Kim, K.H.; Park, K.U.; Kang, E.J.; Choi, Y.H.; Ahn, M.S.; Lee, M.H.; Sun, J.M.; et al. Osimertinib for Patients with Non-Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Multicenter, Open-Label, Phase II Trial (KCSG-LU15-09). J. Clin. Oncol. 2020, 38, 488–495.
- Eide, I.J.Z.; Stensgaard, S.; Helland, A.; Ekman, S.; Mellemgaard, A.; Hansen, K.H.; Cicenas, S.; Koivunen, J.; Gronberg, B.H.; Sorensen, B.S.; et al. Osimertinib in non-small cell lung cancer with uncommon EGFR-mutations: A post-hoc subgroup analysis with pooled data from two phase II clinical trials. Transl. Lung Cancer Res. 2022, 11, 953–963.
- Banno, E.; Togashi, Y.; Nakamura, Y.; Chiba, M.; Kobayashi, Y.; Hayashi, H.; Terashima, M.; de Velasco, M.A.; Sakai, K.; Fujita, Y.; et al. Sensitivities to various epidermal growth factor receptor-tyrosine kinase inhibitors of uncommon epidermal growth factor receptor mutations L861Q and S768I: What is the optimal epidermal growth factor receptor-tyrosine kinase inhibitor? Cancer Sci. 2016, 107, 1134–1140.
- Chiu, C.H.; Yang, C.T.; Shih, J.Y.; Huang, M.S.; Su, W.C.; Lai, R.S.; Wang, C.C.; Hsiao, S.H.; Lin, Y.C.; Ho, C.L.; et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas with G719X/L861Q/S768I Mutations. J. Thorac. Oncol. 2015, 10, 793–799.
- Gallant, J.N.; Sheehan, J.H.; Shaver, T.M.; Bailey, M.; Lipson, D.; Chandramohan, R.; Red Brewer, M.; York, S.J.; Kris, M.G.; Pietenpol, J.A.; et al. EGFR Kinase Domain Duplication (EGFR-KDD) Is a Novel Oncogenic Driver in Lung Cancer That Is Clinically Responsive to Afatinib. Cancer Discov. 2015, 5, 1155–1163.
- Wang, J.; Li, X.; Xue, X.; Ou, Q.; Wu, X.; Liang, Y.; Wang, X.; You, M.; Shao, Y.W.; Zhang, Z.; et al. Clinical outcomes of EGFR kinase domain duplication to targeted therapies in NSCLC. Int. J. Cancer 2019, 144, 2677–2682.
- Costa, D.B. Kinase inhibitor-responsive genotypes in EGFR mutated lung adenocarcinomas: Moving past common point mutations or indels into uncommon kinase domain duplications and rearrangements. Transl. Lung Cancer Res. 2016, 5, 331–337.
- Wu, D.; Xie, Y.; Jin, C.; Qiu, J.; Hou, T.; Du, H.; Chen, S.; Xiang, J.; Shi, X.; Liu, J. The landscape of kinase domain duplication in Chinese lung cancer patients. Ann. Transl. Med. 2020, 8, 1642.
- Baik, C.S.; Wu, D.; Smith, C.; Martins, R.G.; Pritchard, C.C. Durable Response to Tyrosine Kinase Inhibitor Therapy in a Lung Cancer Patient Harboring Epidermal Growth Factor Receptor Tandem Kinase Domain Duplication. J. Thorac. Oncol. 2015, 10, e97-99.
- Chen, D.; Li, X.L.; Wu, B.; Zheng, X.B.; Wang, W.X.; Chen, H.F.; Dong, Y.Y.; Xu, C.W.; Fang, M.Y. A Novel Oncogenic Driver in a Lung Adenocarcinoma Patient Harboring an EGFR-KDD and Response to Afatinib. Front. Oncol. 2020, 10, 867.
- Zhang, L.D.; Gao, H.; Qin, S.M.; Zeng, Q.; Chen, Q.F. Osimertinib is an effective epidermal growth factor receptor-tyrosine kinase inhibitor choice for lung cancer with epidermal growth factor receptor exon 18-25 kinase domain duplication: Report of two cases. Anticancer Drugs 2022, 33, e486–e490.
- Taek Kim, J.; Zhang, W.; Lopategui, J.; Vail, E.; Balmanoukian, A. Patient with Stage IV NSCLC and CNS Metastasis with EGFR Exon 18-25 Kinase Domain Duplication with Response to Osimertinib as a First-Line Therapy. JCO Precis. Oncol. 2021, 5, 88–92.
- Du, Z.; Brown, B.P.; Kim, S.; Ferguson, D.; Pavlick, D.C.; Jayakumaran, G.; Benayed, R.; Gallant, J.N.; Zhang, Y.K.; Yan, Y.; et al. Structure-function analysis of oncogenic EGFR Kinase Domain Duplication reveals insights into activation and a potential approach for therapeutic targeting. Nat. Commun. 2021, 12, 1382.
- Jin, R.; Li, J.; Jin, Z.; Lu, Y.; Shao, Y.W.; Li, W.; Zhao, G.; Xia, Y. Osimertinib confers potent binding affinity to EGFR kinase domain duplication. Int. J. Cancer 2019, 145, 2884–2885.
- Wu, J.Y.; Shih, J.Y. Effectiveness of tyrosine kinase inhibitors on uncommon E709X epidermal growth factor receptor mutations in non-small-cell lung cancer. OncoTargets Ther. 2016, 9, 6137–6145.
- Cheng, C.; Wang, R.; Li, Y.; Pan, Y.; Zhang, Y.; Li, H.; Zheng, D.; Zheng, S.; Shen, X.; Sun, Y.; et al. EGFR Exon 18 Mutations in East Asian Patients with Lung Adenocarcinomas: A Comprehensive Investigation of Prevalence, Clinicopathologic Characteristics and Prognosis. Sci. Rep. 2015, 5, 13959.
