Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Respiratory System
Patients with cystic fibrosis (CF) are repeatedly exposed to antibiotics, especially during the pulmonary exacerbations of the disease. However, the available therapeutic strategies are frequently inadequate to eradicate the involved pathogens and most importantly, facilitate the development of antimicrobial resistance (AMR). The evaluation of AMR is demanding; conventional culture-based susceptibility-testing techniques cannot account for the lung microenvironment and/or the adaptive mechanisms developed by the pathogens, such as biofilm formation.
- cystic fibrosis
- antibiotics
- resistance
1. Background
Cystic fibrosis (CF) is a complex, heterogeneous, multi-organ disorder caused by defects in the CF transmembrane conductance regulator (CFTR) gene. The gene encodes an ion channel that is physiologically involved in the transport of chloride and bicarbonate and is expressed on the apical epithelial cell surfaces throughout the body [1]. CFTR gene defects result in various degrees of ion channel dysfunction, thus ultimately affecting the secretory function of many organs [2]. CF is the most common autosomal recessive life-shortening condition of Caucasians; although its incidence varies significantly, it is estimated that it occurs between 1:3000 and 1:6000 live births, which equates to 1:28 and 1:40 carrier rates, respectively [3].
In the lungs, reduced chloride excretion and unrestricted sodium absorption at the airway epithelial surface result in the overproduction of thick and viscid mucus that affects airway clearance, leading to small airways plugging, irreversible structural and functional defects, and eventually, respiratory failure [4]. Repeated bacterial respiratory infections are the hallmark of the disease [4]; these lead to a domino sequence of persistent lower airway inflammation that accelerates tissue damage and accounts for the majority of the disease-related morbidity and mortality [5]. Common CF pathogens are comprised mainly of Staphylococcus aureus and Pseudomonas aeruginosa; later, as the disease progresses, many patients are infected with more uncommon and difficult-to-treat microorganisms, such as Burkholderia cepacia. Several complications can occur in parallel with the progression of the disease, including liver dysfunction, pancreatic exocrine insufficiency (expressed as malnutrition and failure to thrive in the pediatric population), CF-related diabetes, male infertility, nasal polyposis, intestine obstruction, bronchiectasis, and others [2,6,7].
In individuals with CF, microbial communities colonize the airways shortly after birth, while peripheral lung colonization and chronic inflammation occur later [8]. In contrast to respiratory infections in non-CF patients, these have the tendency to be persistent and involve phenotypic alterations of the infecting organisms [9]. Throughout life, patients with CF are repeatedly treated with antibiotics, especially during the exacerbations of the disease. Antibiotic administration together with novel therapeutic interventions, such as mutation-specific modulator therapies, have substantially expanded life expectancy because the latter relies mainly upon the progression of pulmonary complications [5]. However, accumulating evidence suggests the available therapeutic strategies are frequently inadequate to completely eradicate the involved pathogens [8], while standard dosing schemes may result in sub-therapeutic antibiotic levels, thus increasing the risk of treatment failure and most importantly, augmenting the appearance of antimicrobial resistance (AMR) [10]; the widespread alternating use of ‘suppressive’ and ‘curative’ antibiotic schemes increases the risk of AMR even further [11].
Notwithstanding, the evaluation of AMR is demanding; conventional laboratory antibiotic susceptibility-testing techniques rely on the interpretation of planktonic cultures, which are not entirely representative of the lung (micro) environment [12]. Indeed, the administration of an antibiotic to which a pathogen is resistant in vitro does not steadily prejudge a poor clinical result and vice versa [13]. This is an existing concern in individuals with chronic Pseudomonas aeruginosa and other non-fermenting Gram-negative rod (NFGR) infections [14]. Evidence has also demonstrated pathogens in CF patients commonly form biofilms, which renders them more resistant to a large spectrum of antibiotics [15]. Additionally, features linked to modified pharmacokinetics and pulmonary parenchyma penetration make the dosing of antibiotics in CF even more challenging [16].
2. Antibiotic Susceptibility and Biofilm Formation
CF orchestrates a diversified airway environment due to altered electrolyte levels, the presence of thick mucus, and the release of proteolytic enzymes with subsequent pH decrease; all these features are prosperous towards switching microbial growth from planktonic to biofilm type [8,17] (Figure 1). Biofilms are complex, organized, bacterial structures surrounded by an extracellular polymeric matrix composed of different macromolecules, all building a biophysical barrier that confers pathogens’ protection against the changing surrounding conditions [8,17]. Evidence suggests their formation is cued by quorum-sensing signals, i.e., extracellular chemical signals that control the expression of genes in a cell-density-dependent fashion [18]. In CF, biofilms permit the installation of a microenvironment that promotes maturation and overall shielding of pathogens against both antibiotic actions and host immune mechanisms [17,19]. Biofilm tolerance is rendered to numerous biologic processes, and it escalates as biofilm maturation progresses [19].
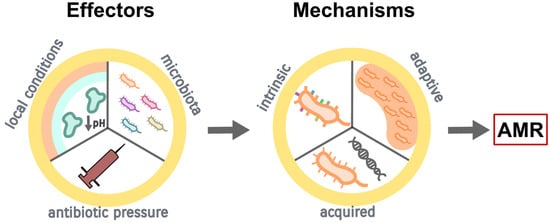
Figure 1. Development of antimicrobial resistance (AMR) in CF (see text for the detailed description).
The presence of biofilms results in the protractedness of lung infections in patients with CF [19], ultimately contributing to the onset of pulmonary exacerbations. Current studies using culture-independent molecular techniques, such as metagenomic sequencing, have shown the complicated dynamics of airway microbiome in CF involve both the ‘traditional’, routinely identified pathogens, as well as other atypical microorganisms [20,21], as discussed below. However, antibiotics are unable to eradicate the biofilm-forming pathogens, firstly, because of their intrinsic antibiotic tolerance and secondly, due to the capacity of biofilms to favor the emergence and spreading of mutational AMR [22]. The mechanism of biofilm tolerance to antibiotics is composite and attributed to physical, physiological, and genetic factors; AMR, on the other hand, is caused by mutations following recurrent exposure to high concentrations of antibiotics [22] (Figure 1).
The formation of biofilms not only protects the bacteria against the host immune system and/or antimicrobial agents but also permits their growth and adaptation in an environment of anoxia and lack of nutrients [8]. Biofilms comprise a remarkable amount of bacterial sub-communities, which are characterized by various degrees of metabolic activity. Peripheral sub-populations demonstrate high metabolic activity, thus consuming large amounts of oxygen and nutrients; on the contrary, sub-populations located at the inner layers have lower or even zero metabolic activity, which renders them more tolerant to antimicrobial agents, thus leading to infection persistence and/or reoccurrence [23]. Higher concentrations of antibiotics or liposomal antibiotic formulations may be more effective, albeit at the price of an increased risk of both toxicity and AMR [24].
Biofilms are currently acknowledged as a fundamental driver of protracted and/or relapsing lung infections in patients with CF. Noteworthy, recently formed biofilms are much more susceptible to antibiotics than the more developed ones [25,26], thus highlighting the need for prompt and well-designed therapeutic strategies.
This entry is adapted from the peer-reviewed paper 10.3390/antibiotics12020217
This entry is offline, you can click here to edit this entry!
